| p-value: | 1e-9 |
| log p-value: | -2.266e+01 |
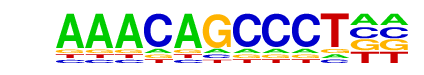
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 46.0 |
| Percentage of Target Sequences with motif | 1.53% |
| Number of Background Sequences with motif | 2.7 |
| Percentage of Background Sequences with motif | 0.11% |
| Average Position of motif in Targets | 37.2 +/- 20.7bp |
| Average Position of motif in Background | 26.5 +/- 13.3bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan_et_al.)/Homer
| Match Rank: | 1 |
| Score: | 0.66
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AAACAGCCCT
GTAAACAG---- |
|


|
|
Foxo1/MA0480.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.66
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---AAACAGCCCT
TGTAAACAGGA-- |
|


|
|
TBF1/MA0403.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.66
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | AAACAGCCCT--
----AACCCTAA |
|

|
|
BMYB(HTH)/Hela-BMYB-ChIP-Seq(GSE27030)/Homer
| Match Rank: | 4 |
| Score: | 0.65
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AAACAGCCCT
NHAACBGYYV- |
|


|
|
mirr/MA0233.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AAACAGCCCT
AAACA----- |
|


|
|
MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer
| Match Rank: | 6 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AAACAGCCCT
YAACBGCC-- |
|


|
|
ara/MA0210.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.63
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AAACAGCCCT
TAACA----- |
|


|
|
Unknown2/Drosophila-Promoters/Homer
| Match Rank: | 8 |
| Score: | 0.63
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | AAACAGCCCT-
---CATCMCTA |
|


|
|
FOXD2/MA0847.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.62
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AAACAGCCCT
GTAAACA----- |
|


|
|
Mirr/dmmpmm(Noyes_hd)/fly
| Match Rank: | 10 |
| Score: | 0.62
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---AAACAGCCCT
AAAAAACAAA--- |
|


|
|





















