| p-value: | 1e-9 |
| log p-value: | -2.264e+01 |
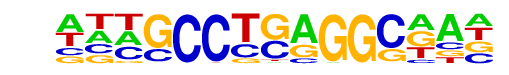
| Information Content per bp: | 1.685 |
| Number of Target Sequences with motif | 37.0 |
| Percentage of Target Sequences with motif | 1.23% |
| Number of Background Sequences with motif | 0.9 |
| Percentage of Background Sequences with motif | 0.04% |
| Average Position of motif in Targets | 44.9 +/- 15.7bp |
| Average Position of motif in Background | 30.4 +/- 8.3bp |
| Strand Bias (log2 ratio + to - strand density) | -0.8 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
TFAP2C(var.2)/MA0814.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.68
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TGCCTGAGACAT
AGCCTCAGGCA- |
|


|
|
TFAP2B(var.2)/MA0812.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.68
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TGCCTGAGACAT
AGCCTCAGGCA- |
|


|
|
PB0190.1_Tcfap2b_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.66
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TGCCTGAGACAT-
ANTGCCTGAGGCAAN |
|


|
|
TFAP2A/MA0003.3/Jaspar
| Match Rank: | 4 |
| Score: | 0.66
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGCCTGAGACAT
NGCCTGAGGCN- |
|


|
|
PB0088.1_Tcfap2e_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.63
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TGCCTGAGACAT-
ATTGCCTGAGGCAAT |
|

|
|
Ap1/dmmpmm(Papatsenko)/fly
| Match Rank: | 6 |
| Score: | 0.61
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | TGCCTGAGACAT
---CTGAGTCAC |
|


|
|
GCN4(MacIsaac)/Yeast
| Match Rank: | 7 |
| Score: | 0.61
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | TGCCTGAGACAT
----TGAGTCAT |
|


|
|
MOT3/Literature(Harbison)/Yeast
| Match Rank: | 8 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGCCTGAGACAT
TACCTN------ |
|


|
|
MOT3(MacIsaac)/Yeast
| Match Rank: | 9 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGCCTGAGACAT
TACCTN------ |
|


|
|
MOT3/MA0340.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGCCTGAGACAT
TACCTN------ |
|


|
|





















