| p-value: | 1e-14 |
| log p-value: | -3.229e+01 |
| Information Content per bp: | 1.729 |
| Number of Target Sequences with motif | 94.0 |
| Percentage of Target Sequences with motif | 19.42% |
| Number of Background Sequences with motif | 12.2 |
| Percentage of Background Sequences with motif | 3.18% |
| Average Position of motif in Targets | 40.0 +/- 18.3bp |
| Average Position of motif in Background | 37.2 +/- 15.8bp |
| Strand Bias (log2 ratio + to - strand density) | 0.4 |
| Multiplicity (# of sites on avg that occur together) | 1.07 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
ASH1/Literature(Harbison)/Yeast
| Match Rank: | 1 |
| Score: | 0.70
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CMTMGTCA-
---AGTCAA |
|


|
|
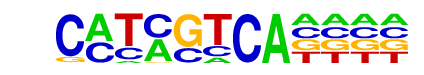
TGA1(bZIP)/Arabidopsis thaliana/AthaMap
| Match Rank: | 2 |
| Score: | 0.68
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CMTMGTCA----
TAACGTCANCAN |
|

|
|
MAFG::NFE2L1/MA0089.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.67
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | CMTMGTCA--
----GTCATN |
|


|
|
WRKY40/MA1085.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.67
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CMTMGTCA--
--TGGTCAAC |
|


|
|
cnc::maf-S/MA0530.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.67
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----CMTMGTCA--
ATNTGCNGAGTCATN |
|


|
|
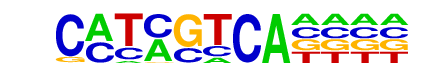
HVH21(HD-KNOTTED)/Hordeum vulgare/AthaMap
| Match Rank: | 6 |
| Score: | 0.66
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CMTMGTCA----
ANCTGTCACNNN |
|

|
|
WRKY48/MA1088.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.66
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CMTMGTCA---
-GAGGTCAACG |
|


|
|
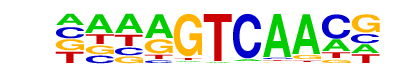
WRKY57/MA1089.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.66
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CMTMGTCA---
-AAAGTCAACG |
|

|
|
Pax2/MA0067.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.66
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CMTMGTCA---
---AGTCACGC |
|


|
|
PB0108.1_Atf1_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.66
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CMTMGTCA---
NTTATTCGTCATNC |
|


|
|





















