| p-value: | 1e-8 |
| log p-value: | -2.059e+01 |
| Information Content per bp: | 1.809 |
| Number of Target Sequences with motif | 47.0 |
| Percentage of Target Sequences with motif | 1.57% |
| Number of Background Sequences with motif | 4.1 |
| Percentage of Background Sequences with motif | 0.15% |
| Average Position of motif in Targets | 37.9 +/- 16.4bp |
| Average Position of motif in Background | 40.4 +/- 14.2bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
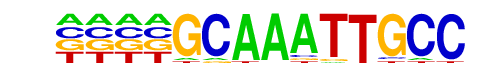
NFkB-p65-Rel(RHD)/ThioMac-LPS-Expression(GSE23622)/Homer
| Match Rank: | 1 |
| Score: | 0.73
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GCAAATTGCC
GGAAATTCCC |
|


|
|
PH0024.1_Dlx5/Jaspar
| Match Rank: | 2 |
| Score: | 0.69
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----GCAAATTGCC--
NANNGNTAATTACCNN |
|


|
|
PH0009.1_Bsx/Jaspar
| Match Rank: | 3 |
| Score: | 0.64
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----GCAAATTGCC--
NTNAGNTAATTACCTN |
|


|
|
RELA/MA0107.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GCAAATTGCC
GGAAATTCCC |
|


|
|
HESX1/MA0894.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.62
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GCAAATTGCC
GCTAATTGGC |
|


|
|
Oct2(POU,Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 6 |
| Score: | 0.62
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GCAAATTGCC
ATATGCAAAT---- |
|

|
|
SD0002.1_at_AC_acceptor/Jaspar
| Match Rank: | 7 |
| Score: | 0.62
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GCAAATTGCC--
-NNACTTGCCTT |
|


|
|
PH0007.1_Barx1/Jaspar
| Match Rank: | 8 |
| Score: | 0.62
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----GCAAATTGCC--
ATNNACTAATTACTTT |
|


|
|
ftz/dmmpmm(Pollard)/fly
| Match Rank: | 9 |
| Score: | 0.61
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | GCAAATTGCC
--TAATTGCC |
|


|
|
GBX2/MA0890.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.61
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GCAAATTGCC
NNTAATTGNN |
|


|
|





















