| p-value: | 1e-9 |
| log p-value: | -2.240e+01 |
| Information Content per bp: | 1.778 |
| Number of Target Sequences with motif | 36.0 |
| Percentage of Target Sequences with motif | 3.85% |
| Number of Background Sequences with motif | 0.8 |
| Percentage of Background Sequences with motif | 0.10% |
| Average Position of motif in Targets | 36.1 +/- 18.5bp |
| Average Position of motif in Background | 45.0 +/- 0.0bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.03 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MBP1::SWI6/MA0330.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.78
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CGCGTTTC
ACGCGTT-- |
|


|
|
STE12/MA0393.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.76
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CGCGTTTC-
--TGTTTCA |
|


|
|
STE12(MacIsaac)/Yeast
| Match Rank: | 3 |
| Score: | 0.76
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CGCGTTTC-
--TGTTTCA |
|


|
|
CMTA3/MA0970.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.75
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CGCGTTTC
CCGCGTTAC |
|


|
|
CMTA2/MA0969.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.75
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CGCGTTTC
AAACCGCGT--- |
|


|
|
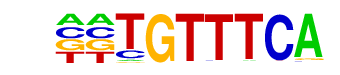
STE12/STE12_Alpha/92-STE12(Harbison)/Yeast
| Match Rank: | 6 |
| Score: | 0.74
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CGCGTTTC-
--TGTTTCA |
|

|
|
MBP1/MA0329.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.72
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CGCGTTTC
NNCGCGT--- |
|


|
|
che-1/MA0260.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.72
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CGCGTTTC
--GGTTTC |
|


|
|
MA0260.1_che-1/Jaspar
| Match Rank: | 9 |
| Score: | 0.72
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CGCGTTTC
--GGTTTC |
|


|
|
DIG1/DIG1_YPD/32-STE12(Harbison)/Yeast
| Match Rank: | 10 |
| Score: | 0.72
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CGCGTTTC-
--TGTTTCA |
|


|
|





















