| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | PHA-4(Forkhead)/cElegans-Embryos-PHA4-ChIP-Seq(modEncode)/Homer | 1e-44 | -1.030e+02 | 0.0000 | 418.0 | 44.75% | 112.0 | 14.15% | motif file (matrix) |
pdf |
| 2 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-28 | -6.450e+01 | 0.0000 | 237.0 | 25.37% | 49.8 | 6.29% | motif file (matrix) |
pdf |
| 3 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-27 | -6.325e+01 | 0.0000 | 155.0 | 16.60% | 15.6 | 1.97% | motif file (matrix) |
pdf |
| 4 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-24 | -5.729e+01 | 0.0000 | 209.0 | 22.38% | 42.8 | 5.41% | motif file (matrix) |
pdf |
| 5 |  | FOXM1(Forkhead)/MCF7-FOXM1-ChIP-Seq(GSE72977)/Homer | 1e-23 | -5.457e+01 | 0.0000 | 210.0 | 22.48% | 45.0 | 5.69% | motif file (matrix) |
pdf |
| 6 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan_et_al.)/Homer | 1e-17 | -3.931e+01 | 0.0000 | 207.0 | 22.16% | 60.8 | 7.68% | motif file (matrix) |
pdf |
| 7 |  | FoxL2(Forkhead)/Ovary-FoxL2-ChIP-Seq(GSE60858)/Homer | 1e-16 | -3.867e+01 | 0.0000 | 147.0 | 15.74% | 31.0 | 3.91% | motif file (matrix) |
pdf |
| 8 |  | Fox:Ebox(Forkhead,bHLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-15 | -3.468e+01 | 0.0000 | 128.0 | 13.70% | 25.2 | 3.19% | motif file (matrix) |
pdf |
| 9 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-14 | -3.435e+01 | 0.0000 | 116.0 | 12.42% | 20.6 | 2.60% | motif file (matrix) |
pdf |
| 10 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-14 | -3.373e+01 | 0.0000 | 95.0 | 10.17% | 13.0 | 1.64% | motif file (matrix) |
pdf |
| 11 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-12 | -2.850e+01 | 0.0000 | 95.0 | 10.17% | 16.7 | 2.11% | motif file (matrix) |
pdf |
| 12 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-12 | -2.837e+01 | 0.0000 | 181.0 | 19.38% | 60.6 | 7.66% | motif file (matrix) |
pdf |
| 13 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-10 | -2.429e+01 | 0.0000 | 116.0 | 12.42% | 30.3 | 3.82% | motif file (matrix) |
pdf |
| 14 |  | TEAD2(TEA)/Py2T-Tead2-ChIP-Seq(GSE55709)/Homer | 1e-10 | -2.354e+01 | 0.0000 | 73.0 | 7.82% | 11.3 | 1.43% | motif file (matrix) |
pdf |
| 15 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-8 | -2.067e+01 | 0.0000 | 81.0 | 8.67% | 17.5 | 2.21% | motif file (matrix) |
pdf |
| 16 |  | NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer | 1e-6 | -1.602e+01 | 0.0000 | 153.0 | 16.38% | 64.0 | 8.09% | motif file (matrix) |
pdf |
| 17 | 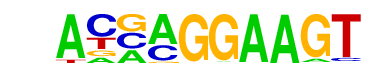 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-5 | -1.286e+01 | 0.0001 | 57.0 | 6.10% | 14.9 | 1.88% | motif file (matrix) |
pdf |
| 18 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-5 | -1.205e+01 | 0.0001 | 61.0 | 6.53% | 18.0 | 2.27% | motif file (matrix) |
pdf |
| 19 |  | Etv2(ETS)/ES-ER71-ChIP-Seq(GSE59402)/Homer(0.967) | 1e-5 | -1.168e+01 | 0.0002 | 48.0 | 5.14% | 11.5 | 1.46% | motif file (matrix) |
pdf |
| 20 | 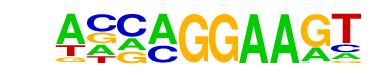 | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-5 | -1.155e+01 | 0.0002 | 85.0 | 9.10% | 31.4 | 3.97% | motif file (matrix) |
pdf |
| 21 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-4 | -1.140e+01 | 0.0002 | 93.0 | 9.96% | 36.7 | 4.64% | motif file (matrix) |
pdf |
| 22 |  | Stat3+il21(Stat)/CD4-Stat3-ChIP-Seq(GSE19198)/Homer | 1e-4 | -1.082e+01 | 0.0004 | 54.0 | 5.78% | 15.7 | 1.99% | motif file (matrix) |
pdf |
| 23 |  | STAT4(Stat)/CD4-Stat4-ChIP-Seq(GSE22104)/Homer | 1e-4 | -1.029e+01 | 0.0006 | 62.0 | 6.64% | 20.7 | 2.62% | motif file (matrix) |
pdf |
| 24 |  | GATA(Zf),IR4/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-4 | -9.255e+00 | 0.0015 | 15.0 | 1.61% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 25 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-3 | -9.077e+00 | 0.0018 | 29.0 | 3.10% | 5.6 | 0.71% | motif file (matrix) |
pdf |
| 26 |  | PQM-1(?)/cElegans-L3-ChIP-Seq(modEncode)/Homer | 1e-3 | -9.039e+00 | 0.0018 | 24.0 | 2.57% | 3.3 | 0.42% | motif file (matrix) |
pdf |
| 27 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-3 | -8.801e+00 | 0.0022 | 45.0 | 4.82% | 13.7 | 1.73% | motif file (matrix) |
pdf |
| 28 |  | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-3 | -8.788e+00 | 0.0022 | 56.0 | 6.00% | 19.3 | 2.44% | motif file (matrix) |
pdf |
| 29 |  | Fosl2(bZIP)/3T3L1-Fosl2-ChIP-Seq(GSE56872)/Homer | 1e-3 | -8.780e+00 | 0.0022 | 26.0 | 2.78% | 4.4 | 0.56% | motif file (matrix) |
pdf |
| 30 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-3 | -8.462e+00 | 0.0027 | 46.0 | 4.93% | 14.9 | 1.88% | motif file (matrix) |
pdf |
| 31 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-3 | -8.363e+00 | 0.0029 | 60.0 | 6.42% | 22.3 | 2.82% | motif file (matrix) |
pdf |
| 32 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.349e+00 | 0.0029 | 20.0 | 2.14% | 2.1 | 0.27% | motif file (matrix) |
pdf |
| 33 | 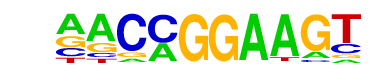 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-3 | -8.134e+00 | 0.0034 | 71.0 | 7.60% | 29.7 | 3.75% | motif file (matrix) |
pdf |
| 34 |  | LHY(Myb)/Seedling-LHY-ChIP-Seq(GSE52175)/Homer | 1e-3 | -8.118e+00 | 0.0034 | 66.0 | 7.07% | 26.7 | 3.38% | motif file (matrix) |
pdf |
| 35 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-3 | -7.846e+00 | 0.0043 | 55.0 | 5.89% | 20.7 | 2.62% | motif file (matrix) |
pdf |
| 36 |  | caudal(Homeobox)/Drosophila-Embryos-ChIP-Chip(modEncode)/Homer | 1e-3 | -7.756e+00 | 0.0046 | 53.0 | 5.67% | 19.9 | 2.52% | motif file (matrix) |
pdf |
| 37 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.541e+00 | 0.0056 | 30.0 | 3.21% | 7.5 | 0.95% | motif file (matrix) |
pdf |
| 38 |  | Sox4(HMG)/proB-Sox4-ChIP-Seq(GSE50066)/Homer | 1e-3 | -7.003e+00 | 0.0093 | 42.0 | 4.50% | 14.8 | 1.87% | motif file (matrix) |
pdf |
| 39 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-2 | -6.763e+00 | 0.0115 | 34.0 | 3.64% | 10.2 | 1.29% | motif file (matrix) |
pdf |
| 40 |  | PABPC1(?)/MEL-PABC1-CLIP-Seq(GSE69755)/Homer | 1e-2 | -6.745e+00 | 0.0115 | 155.0 | 16.60% | 90.2 | 11.39% | motif file (matrix) |
pdf |
| 41 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-2 | -6.652e+00 | 0.0122 | 41.0 | 4.39% | 14.6 | 1.84% | motif file (matrix) |
pdf |
| 42 |  | Tal1 | 1e-2 | -6.546e+00 | 0.0132 | 81.0 | 8.67% | 39.5 | 4.99% | motif file (matrix) |
pdf |
| 43 |  | HOXB13(Homeobox)/ProstateTumor-HOXB13-ChIP-Seq(GSE56288)/Homer | 1e-2 | -6.541e+00 | 0.0132 | 84.0 | 8.99% | 41.1 | 5.20% | motif file (matrix) |
pdf |
| 44 |  | BMYB(HTH)/Hela-BMYB-ChIP-Seq(GSE27030)/Homer | 1e-2 | -6.417e+00 | 0.0144 | 85.0 | 9.10% | 42.8 | 5.41% | motif file (matrix) |
pdf |
| 45 |  | ELT-3(Gata)/cElegans-L1-ELT3-ChIP-Seq(modEncode)/Homer | 1e-2 | -6.384e+00 | 0.0145 | 33.0 | 3.53% | 10.5 | 1.33% | motif file (matrix) |
pdf |
| 46 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-2 | -6.286e+00 | 0.0157 | 27.0 | 2.89% | 7.5 | 0.95% | motif file (matrix) |
pdf |
| 47 |  | GRHL2(CP2)/HBE-GRHL2-ChIP-Seq(GSE46194)/Homer | 1e-2 | -5.911e+00 | 0.0223 | 28.0 | 3.00% | 8.9 | 1.13% | motif file (matrix) |
pdf |
| 48 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-2 | -5.830e+00 | 0.0237 | 42.0 | 4.50% | 16.5 | 2.08% | motif file (matrix) |
pdf |
| 49 |  | SCL(bHLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-2 | -5.772e+00 | 0.0246 | 224.0 | 23.98% | 146.2 | 18.47% | motif file (matrix) |
pdf |
| 50 |  | Oct4(POU,Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.541e+00 | 0.0303 | 36.0 | 3.85% | 13.2 | 1.66% | motif file (matrix) |
pdf |
| 51 |  | ZNF467(Zf)/HEK293-ZNF467.GFP-ChIP-Seq(GSE58341)/Homer | 1e-2 | -5.458e+00 | 0.0323 | 23.0 | 2.46% | 6.7 | 0.85% | motif file (matrix) |
pdf |
| 52 |  | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer | 1e-2 | -5.347e+00 | 0.0355 | 47.0 | 5.03% | 20.1 | 2.54% | motif file (matrix) |
pdf |
| 53 |  | SeqBias: polyA-repeat | 1e-2 | -5.094e+00 | 0.0448 | 892.0 | 95.50% | 732.2 | 92.50% | motif file (matrix) |
pdf |
| 54 |  | Oct2(POU,Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-2 | -5.054e+00 | 0.0458 | 22.0 | 2.36% | 6.6 | 0.84% | motif file (matrix) |
pdf |
| 55 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.046e+00 | 0.0458 | 18.0 | 1.93% | 4.3 | 0.55% | motif file (matrix) |
pdf |
| 56 |  | E2A(bHLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-2 | -4.983e+00 | 0.0474 | 52.0 | 5.57% | 24.7 | 3.12% | motif file (matrix) |
pdf |
| 57 |  | Zfp809(Zf)/ES-Zfp809-ChIP-Seq(GSE70799)/Homer | 1e-2 | -4.922e+00 | 0.0495 | 8.0 | 0.86% | 0.4 | 0.05% | motif file (matrix) |
pdf |
| 58 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-2 | -4.785e+00 | 0.0557 | 37.0 | 3.96% | 15.4 | 1.94% | motif file (matrix) |
pdf |
| 59 |  | Zelda(Zf)/Embryo-zld-ChIP-Seq(GSE65441)/Homer | 1e-2 | -4.622e+00 | 0.0645 | 19.0 | 2.03% | 5.7 | 0.72% | motif file (matrix) |
pdf |
| 60 |  | ETS(ETS)/Promoter/Homer | 1e-2 | -4.612e+00 | 0.0645 | 17.0 | 1.82% | 4.9 | 0.62% | motif file (matrix) |
pdf |























































































































