


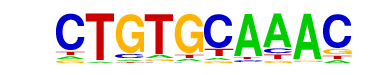


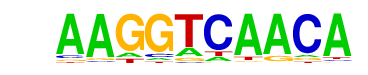



| p-value: | 1e-165 |
| log p-value: | -3.819e+02 |
| Information Content per bp: | 1.743 |
| Number of Target Sequences with motif | 1492.0 |
| Percentage of Target Sequences with motif | 49.73% |
| Number of Background Sequences with motif | 432.5 |
| Percentage of Background Sequences with motif | 16.00% |
| Average Position of motif in Targets | 36.3 +/- 20.6bp |
| Average Position of motif in Background | 35.6 +/- 24.1bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.34 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.953 |  | 1e-151 | -348.763878 | 51.63% | 18.78% | motif file (matrix) |
| 2 | 0.950 |  | 1e-120 | -278.370377 | 45.33% | 16.89% | motif file (matrix) |
| 3 | 0.830 |  | 1e-40 | -93.743340 | 15.27% | 4.74% | motif file (matrix) |
| 4 | 0.627 |  | 1e-20 | -48.243361 | 4.07% | 0.51% | motif file (matrix) |
| 5 | 0.661 |  | 1e-15 | -35.989696 | 3.23% | 0.48% | motif file (matrix) |
| 6 | 0.632 |  | 1e-14 | -32.706873 | 2.00% | 0.09% | motif file (matrix) |
| 7 | 0.604 |  | 1e-10 | -24.233849 | 2.57% | 0.51% | motif file (matrix) |
| 8 | 0.618 |  | 1e-10 | -23.497547 | 4.70% | 1.74% | motif file (matrix) |