



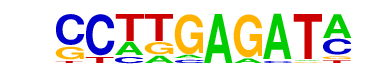





| p-value: | 1e-97 |
| log p-value: | -2.242e+02 |
| Information Content per bp: | 1.810 |
| Number of Target Sequences with motif | 965.0 |
| Percentage of Target Sequences with motif | 32.17% |
| Number of Background Sequences with motif | 266.6 |
| Percentage of Background Sequences with motif | 9.87% |
| Average Position of motif in Targets | 38.1 +/- 20.7bp |
| Average Position of motif in Background | 38.2 +/- 23.3bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.09 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.761 |  | 1e-55 | -127.833120 | 16.93% | 4.36% | motif file (matrix) |
| 2 | 0.898 |  | 1e-54 | -125.186561 | 12.30% | 2.04% | motif file (matrix) |
| 3 | 0.647 |  | 1e-42 | -98.344182 | 7.33% | 0.66% | motif file (matrix) |
| 4 | 0.668 |  | 1e-34 | -80.231319 | 4.80% | 0.18% | motif file (matrix) |
| 5 | 0.666 |  | 1e-19 | -45.502957 | 4.60% | 0.80% | motif file (matrix) |
| 6 | 0.653 |  | 1e-18 | -43.657562 | 5.90% | 1.49% | motif file (matrix) |
| 7 | 0.742 |  | 1e-16 | -37.585004 | 4.47% | 0.98% | motif file (matrix) |
| 8 | 0.657 |  | 1e-9 | -22.907754 | 3.60% | 1.08% | motif file (matrix) |