| p-value: | 1e-49 |
| log p-value: | -1.142e+02 |
| Information Content per bp: | 1.799 |
| Number of Target Sequences with motif | 833.0 |
| Percentage of Target Sequences with motif | 27.77% |
| Number of Background Sequences with motif | 327.1 |
| Percentage of Background Sequences with motif | 12.10% |
| Average Position of motif in Targets | 37.3 +/- 20.0bp |
| Average Position of motif in Background | 38.5 +/- 24.7bp |
| Strand Bias (log2 ratio + to - strand density) | 0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.10 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer
| Match Rank: | 1 |
| Score: | 0.92
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CAGGAAGT-
ACAGGAAGTG |
|


|
|
Eip74EF/dmmpmm(Bigfoot)/fly
| Match Rank: | 2 |
| Score: | 0.91
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CAGGAAGT
CAGGAAGT |
|


|
|
EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 3 |
| Score: | 0.89
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CAGGAAGT
NACAGGAAAT |
|


|
|
EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 4 |
| Score: | 0.88
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CAGGAAGT
NACAGGAAAT |
|


|
|
Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer
| Match Rank: | 5 |
| Score: | 0.87
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CAGGAAGT
AACAGGAAGT |
|


|
|
Eip74EF/dmmpmm(Pollard)/fly
| Match Rank: | 6 |
| Score: | 0.87
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CAGGAAGT
AACAGGAAGT |
|


|
|
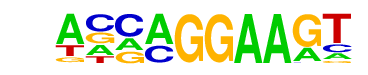
EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer
| Match Rank: | 7 |
| Score: | 0.87
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CAGGAAGT
AVCAGGAAGT |
|

|
|
ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer
| Match Rank: | 8 |
| Score: | 0.86
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CAGGAAGT-
ACAGGAAGTG |
|


|
|
Etv2(ETS)/ES-ER71-ChIP-Seq(GSE59402)/Homer(0.967)
| Match Rank: | 9 |
| Score: | 0.85
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CAGGAAGT--
NDCAGGAARTNN |
|


|
|
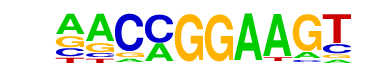
ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer
| Match Rank: | 10 |
| Score: | 0.81
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CAGGAAGT
AACCGGAAGT |
|

|
|





















