| p-value: | 1e-21 |
| log p-value: | -4.879e+01 |
| Information Content per bp: | 1.606 |
| Number of Target Sequences with motif | 351.0 |
| Percentage of Target Sequences with motif | 11.70% |
| Number of Background Sequences with motif | 129.9 |
| Percentage of Background Sequences with motif | 4.81% |
| Average Position of motif in Targets | 37.3 +/- 19.0bp |
| Average Position of motif in Background | 33.5 +/- 17.1bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.05 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
TFAP2B/MA0811.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.85
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CCCTRGGGCW
TGCCCCAGGGCA |
|


|
|
TFAP2C/MA0524.2/Jaspar
| Match Rank: | 2 |
| Score: | 0.85
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CCCTRGGGCW
TGCCCCAGGGCA |
|


|
|
TFAP2A(var.2)/MA0810.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.85
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CCCTRGGGCW
TGCCCCCGGGCA |
|


|
|
PB0086.1_Tcfap2b_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.83
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CCCTRGGGCW-
TTGCCCTAGGGCAT |
|


|
|
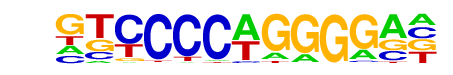
EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 5 |
| Score: | 0.82
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CCCTRGGGCW
GTCCCCAGGGGA- |
|

|
|
EBF(EBF)/proBcell-EBF-ChIP-Seq(GSE21978)/Homer
| Match Rank: | 6 |
| Score: | 0.77
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CCCTRGGGCW
NGTCCCNNGGGA- |
|


|
|
AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 7 |
| Score: | 0.77
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CCCTRGGGCW
ATGCCCTGAGGC- |
|


|
|
EBF1/MA0154.3/Jaspar
| Match Rank: | 8 |
| Score: | 0.76
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CCCTRGGGCW-
ANTCCCTNGGGAAT |
|


|
|
AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer
| Match Rank: | 9 |
| Score: | 0.76
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CCCTRGGGCW-
SCCTSAGGSCAW |
|


|
|
PB0191.1_Tcfap2c_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.76
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CCCTRGGGCW-
CCGCCCAAGGGCAG |
|


|
|





















