| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | PHA-4(Forkhead)/cElegans-Embryos-PHA4-ChIP-Seq(modEncode)/Homer | 1e-123 | -2.839e+02 | 0.0000 | 1327.0 | 44.23% | 427.1 | 15.81% | motif file (matrix) |
pdf |
| 2 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-88 | -2.043e+02 | 0.0000 | 775.0 | 25.83% | 180.3 | 6.67% | motif file (matrix) |
pdf |
| 3 |  | FOXM1(Forkhead)/MCF7-FOXM1-ChIP-Seq(GSE72977)/Homer | 1e-85 | -1.977e+02 | 0.0000 | 673.0 | 22.43% | 134.6 | 4.98% | motif file (matrix) |
pdf |
| 4 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-80 | -1.860e+02 | 0.0000 | 674.0 | 22.47% | 145.7 | 5.39% | motif file (matrix) |
pdf |
| 5 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-69 | -1.593e+02 | 0.0000 | 778.0 | 25.93% | 230.7 | 8.54% | motif file (matrix) |
pdf |
| 6 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-66 | -1.521e+02 | 0.0000 | 513.0 | 17.10% | 97.5 | 3.61% | motif file (matrix) |
pdf |
| 7 |  | Fox:Ebox(Forkhead,bHLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-56 | -1.309e+02 | 0.0000 | 467.0 | 15.57% | 94.3 | 3.49% | motif file (matrix) |
pdf |
| 8 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-51 | -1.178e+02 | 0.0000 | 541.0 | 18.03% | 145.3 | 5.38% | motif file (matrix) |
pdf |
| 9 |  | FoxL2(Forkhead)/Ovary-FoxL2-ChIP-Seq(GSE60858)/Homer | 1e-48 | -1.127e+02 | 0.0000 | 456.0 | 15.20% | 107.0 | 3.96% | motif file (matrix) |
pdf |
| 10 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-46 | -1.065e+02 | 0.0000 | 301.0 | 10.03% | 41.5 | 1.53% | motif file (matrix) |
pdf |
| 11 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan_et_al.)/Homer | 1e-42 | -9.695e+01 | 0.0000 | 643.0 | 21.43% | 232.9 | 8.62% | motif file (matrix) |
pdf |
| 12 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-39 | -9.100e+01 | 0.0000 | 349.0 | 11.63% | 75.8 | 2.81% | motif file (matrix) |
pdf |
| 13 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-34 | -7.925e+01 | 0.0000 | 370.0 | 12.33% | 98.3 | 3.64% | motif file (matrix) |
pdf |
| 14 |  | GATA(Zf),IR3/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-31 | -7.301e+01 | 0.0000 | 180.0 | 6.00% | 18.9 | 0.70% | motif file (matrix) |
pdf |
| 15 |  | GATA(Zf),IR4/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-29 | -6.714e+01 | 0.0000 | 142.0 | 4.73% | 9.3 | 0.34% | motif file (matrix) |
pdf |
| 16 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-28 | -6.668e+01 | 0.0000 | 282.0 | 9.40% | 68.0 | 2.51% | motif file (matrix) |
pdf |
| 17 |  | TEAD2(TEA)/Py2T-Tead2-ChIP-Seq(GSE55709)/Homer | 1e-27 | -6.440e+01 | 0.0000 | 222.0 | 7.40% | 41.3 | 1.53% | motif file (matrix) |
pdf |
| 18 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-27 | -6.269e+01 | 0.0000 | 325.0 | 10.83% | 94.1 | 3.48% | motif file (matrix) |
pdf |
| 19 |  | GATA3(Zf),DR8/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-21 | -4.901e+01 | 0.0000 | 98.0 | 3.27% | 5.0 | 0.19% | motif file (matrix) |
pdf |
| 20 |  | NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer | 1e-16 | -3.838e+01 | 0.0000 | 442.0 | 14.73% | 208.0 | 7.70% | motif file (matrix) |
pdf |
| 21 |  | GRHL2(CP2)/HBE-GRHL2-ChIP-Seq(GSE46194)/Homer | 1e-16 | -3.748e+01 | 0.0000 | 136.0 | 4.53% | 27.9 | 1.03% | motif file (matrix) |
pdf |
| 22 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-14 | -3.375e+01 | 0.0000 | 295.0 | 9.83% | 122.0 | 4.51% | motif file (matrix) |
pdf |
| 23 |  | GATA3(Zf),DR4/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-12 | -2.988e+01 | 0.0000 | 69.0 | 2.30% | 6.1 | 0.23% | motif file (matrix) |
pdf |
| 24 |  | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer | 1e-12 | -2.813e+01 | 0.0000 | 202.0 | 6.73% | 74.4 | 2.75% | motif file (matrix) |
pdf |
| 25 |  | AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer | 1e-11 | -2.743e+01 | 0.0000 | 162.0 | 5.40% | 52.9 | 1.96% | motif file (matrix) |
pdf |
| 26 |  | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-11 | -2.738e+01 | 0.0000 | 176.0 | 5.87% | 60.4 | 2.24% | motif file (matrix) |
pdf |
| 27 |  | PQM-1(?)/cElegans-L3-ChIP-Seq(modEncode)/Homer | 1e-11 | -2.552e+01 | 0.0000 | 157.0 | 5.23% | 52.6 | 1.95% | motif file (matrix) |
pdf |
| 28 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-10 | -2.442e+01 | 0.0000 | 147.0 | 4.90% | 48.2 | 1.78% | motif file (matrix) |
pdf |
| 29 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-10 | -2.418e+01 | 0.0000 | 316.0 | 10.53% | 156.7 | 5.80% | motif file (matrix) |
pdf |
| 30 |  | Fosl2(bZIP)/3T3L1-Fosl2-ChIP-Seq(GSE56872)/Homer | 1e-10 | -2.331e+01 | 0.0000 | 82.0 | 2.73% | 16.3 | 0.60% | motif file (matrix) |
pdf |
| 31 |  | PABPC1(?)/MEL-PABC1-CLIP-Seq(GSE69755)/Homer | 1e-9 | -2.261e+01 | 0.0000 | 422.0 | 14.07% | 236.0 | 8.73% | motif file (matrix) |
pdf |
| 32 |  | HOXB13(Homeobox)/ProstateTumor-HOXB13-ChIP-Seq(GSE56288)/Homer | 1e-8 | -2.003e+01 | 0.0000 | 248.0 | 8.27% | 120.4 | 4.46% | motif file (matrix) |
pdf |
| 33 |  | Fra1(bZIP)/BT549-Fra1-ChIP-Seq(GSE46166)/Homer | 1e-8 | -1.965e+01 | 0.0000 | 113.0 | 3.77% | 36.7 | 1.36% | motif file (matrix) |
pdf |
| 34 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-8 | -1.916e+01 | 0.0000 | 136.0 | 4.53% | 50.5 | 1.87% | motif file (matrix) |
pdf |
| 35 |  | ELT-3(Gata)/cElegans-L1-ELT3-ChIP-Seq(modEncode)/Homer | 1e-8 | -1.874e+01 | 0.0000 | 161.0 | 5.37% | 66.4 | 2.46% | motif file (matrix) |
pdf |
| 36 |  | Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer | 1e-7 | -1.790e+01 | 0.0000 | 218.0 | 7.27% | 105.4 | 3.90% | motif file (matrix) |
pdf |
| 37 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-7 | -1.698e+01 | 0.0000 | 62.0 | 2.07% | 13.2 | 0.49% | motif file (matrix) |
pdf |
| 38 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-7 | -1.674e+01 | 0.0000 | 221.0 | 7.37% | 110.8 | 4.10% | motif file (matrix) |
pdf |
| 39 |  | SeqBias: polyA-repeat | 1e-7 | -1.620e+01 | 0.0000 | 2910.0 | 97.00% | 2544.4 | 94.15% | motif file (matrix) |
pdf |
| 40 |  | Stat3+il21(Stat)/CD4-Stat3-ChIP-Seq(GSE19198)/Homer | 1e-6 | -1.464e+01 | 0.0000 | 132.0 | 4.40% | 57.0 | 2.11% | motif file (matrix) |
pdf |
| 41 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-6 | -1.449e+01 | 0.0000 | 144.0 | 4.80% | 64.3 | 2.38% | motif file (matrix) |
pdf |
| 42 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-6 | -1.387e+01 | 0.0000 | 120.0 | 4.00% | 50.9 | 1.88% | motif file (matrix) |
pdf |
| 43 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-5 | -1.380e+01 | 0.0000 | 55.0 | 1.83% | 13.7 | 0.51% | motif file (matrix) |
pdf |
| 44 |  | Tal1 | 1e-5 | -1.331e+01 | 0.0000 | 314.0 | 10.47% | 188.7 | 6.98% | motif file (matrix) |
pdf |
| 45 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-5 | -1.301e+01 | 0.0000 | 47.0 | 1.57% | 10.8 | 0.40% | motif file (matrix) |
pdf |
| 46 |  | Unknown5/Drosophila-Promoters/Homer | 1e-5 | -1.266e+01 | 0.0000 | 130.0 | 4.33% | 59.6 | 2.20% | motif file (matrix) |
pdf |
| 47 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-4 | -1.131e+01 | 0.0001 | 159.0 | 5.30% | 82.9 | 3.07% | motif file (matrix) |
pdf |
| 48 |  | E-box/Arabidopsis-Promoters/Homer | 1e-4 | -1.074e+01 | 0.0002 | 44.0 | 1.47% | 11.2 | 0.42% | motif file (matrix) |
pdf |
| 49 |  | NF1:FOXA1(CTF,Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-4 | -1.052e+01 | 0.0002 | 33.0 | 1.10% | 6.8 | 0.25% | motif file (matrix) |
pdf |
| 50 |  | Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer | 1e-4 | -1.026e+01 | 0.0003 | 94.0 | 3.13% | 41.1 | 1.52% | motif file (matrix) |
pdf |
| 51 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-4 | -9.656e+00 | 0.0005 | 119.0 | 3.97% | 59.5 | 2.20% | motif file (matrix) |
pdf |
| 52 |  | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-4 | -9.575e+00 | 0.0005 | 252.0 | 8.40% | 156.8 | 5.80% | motif file (matrix) |
pdf |
| 53 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-4 | -9.525e+00 | 0.0005 | 143.0 | 4.77% | 76.4 | 2.83% | motif file (matrix) |
pdf |
| 54 |  | NF1(CTF)/LNCAP-NF1-ChIP-Seq(Unpublished)/Homer | 1e-3 | -8.906e+00 | 0.0010 | 69.0 | 2.30% | 28.9 | 1.07% | motif file (matrix) |
pdf |
| 55 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-3 | -8.728e+00 | 0.0011 | 170.0 | 5.67% | 98.0 | 3.63% | motif file (matrix) |
pdf |
| 56 |  | Esrrb(NR)/mES-Esrrb-ChIP-Seq(GSE11431)/Homer | 1e-3 | -8.273e+00 | 0.0018 | 81.0 | 2.70% | 37.1 | 1.37% | motif file (matrix) |
pdf |
| 57 |  | Sox4(HMG)/proB-Sox4-ChIP-Seq(GSE50066)/Homer | 1e-3 | -7.985e+00 | 0.0023 | 139.0 | 4.63% | 78.2 | 2.89% | motif file (matrix) |
pdf |
| 58 |  | Unknown3/Arabidopsis-Promoters/Homer | 1e-3 | -7.819e+00 | 0.0027 | 23.0 | 0.77% | 4.1 | 0.15% | motif file (matrix) |
pdf |
| 59 |  | Oct4(POU,Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-3 | -7.814e+00 | 0.0027 | 113.0 | 3.77% | 60.5 | 2.24% | motif file (matrix) |
pdf |
| 60 |  | Sp1(Zf)/Promoter/Homer | 1e-3 | -7.709e+00 | 0.0029 | 18.0 | 0.60% | 2.1 | 0.08% | motif file (matrix) |
pdf |
| 61 |  | ZNF416(Zf)/HEK293-ZNF416.GFP-ChIP-Seq(GSE58341)/Homer | 1e-3 | -7.647e+00 | 0.0030 | 186.0 | 6.20% | 114.6 | 4.24% | motif file (matrix) |
pdf |
| 62 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.555e+00 | 0.0033 | 15.0 | 0.50% | 1.1 | 0.04% | motif file (matrix) |
pdf |
| 63 |  | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer | 1e-3 | -7.390e+00 | 0.0038 | 114.0 | 3.80% | 62.1 | 2.30% | motif file (matrix) |
pdf |
| 64 |  | LHY(Myb)/Seedling-LHY-ChIP-Seq(GSE52175)/Homer | 1e-3 | -7.229e+00 | 0.0044 | 228.0 | 7.60% | 148.9 | 5.51% | motif file (matrix) |
pdf |
| 65 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer | 1e-3 | -7.196e+00 | 0.0045 | 65.0 | 2.17% | 29.7 | 1.10% | motif file (matrix) |
pdf |
| 66 |  | Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.163e+00 | 0.0045 | 17.0 | 0.57% | 2.9 | 0.11% | motif file (matrix) |
pdf |
| 67 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-3 | -6.971e+00 | 0.0054 | 14.0 | 0.47% | 1.1 | 0.04% | motif file (matrix) |
pdf |
| 68 |  | CLOCK(bHLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-3 | -6.946e+00 | 0.0055 | 50.0 | 1.67% | 20.7 | 0.77% | motif file (matrix) |
pdf |
| 69 |  | SeqBias: A/T bias | 1e-3 | -6.920e+00 | 0.0055 | 2386.0 | 79.53% | 2056.0 | 76.08% | motif file (matrix) |
pdf |
| 70 |  | STAT4(Stat)/CD4-Stat4-ChIP-Seq(GSE22104)/Homer | 1e-2 | -6.888e+00 | 0.0056 | 158.0 | 5.27% | 96.4 | 3.57% | motif file (matrix) |
pdf |
| 71 |  | Etv2(ETS)/ES-ER71-ChIP-Seq(GSE59402)/Homer(0.967) | 1e-2 | -6.834e+00 | 0.0059 | 167.0 | 5.57% | 103.3 | 3.82% | motif file (matrix) |
pdf |
| 72 | 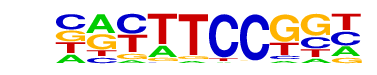 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-2 | -6.695e+00 | 0.0066 | 182.0 | 6.07% | 116.0 | 4.29% | motif file (matrix) |
pdf |
| 73 |  | HLH-1(bHLH)/cElegans-Embryo-HLH1-ChIP-Seq(modEncode)/Homer | 1e-2 | -6.617e+00 | 0.0071 | 127.0 | 4.23% | 74.6 | 2.76% | motif file (matrix) |
pdf |
| 74 |  | Unknown2/Drosophila-Promoters/Homer | 1e-2 | -6.458e+00 | 0.0082 | 157.0 | 5.23% | 97.4 | 3.60% | motif file (matrix) |
pdf |
| 75 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-2 | -6.452e+00 | 0.0082 | 107.0 | 3.57% | 60.1 | 2.22% | motif file (matrix) |
pdf |
| 76 |  | Pho4(bHLH)/Yeast-Pho4-ChIP-Seq(GSE29506)/Homer | 1e-2 | -6.358e+00 | 0.0088 | 20.0 | 0.67% | 4.1 | 0.15% | motif file (matrix) |
pdf |
| 77 |  | Sox10(HMG)/SciaticNerve-Sox3-ChIP-Seq(GSE35132)/Homer | 1e-2 | -6.221e+00 | 0.0100 | 262.0 | 8.73% | 180.0 | 6.66% | motif file (matrix) |
pdf |
| 78 |  | STAT1(Stat)/HelaS3-STAT1-ChIP-Seq(GSE12782)/Homer | 1e-2 | -6.091e+00 | 0.0112 | 52.0 | 1.73% | 24.0 | 0.89% | motif file (matrix) |
pdf |
| 79 |  | IBL1(bHLH)/Seedling-IBL1-ChIP-Seq(GSE51120)/Homer | 1e-2 | -6.043e+00 | 0.0116 | 156.0 | 5.20% | 98.3 | 3.64% | motif file (matrix) |
pdf |
| 80 |  | BMYB(HTH)/Hela-BMYB-ChIP-Seq(GSE27030)/Homer | 1e-2 | -5.983e+00 | 0.0122 | 244.0 | 8.13% | 167.2 | 6.19% | motif file (matrix) |
pdf |
| 81 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-2 | -5.927e+00 | 0.0127 | 78.0 | 2.60% | 41.9 | 1.55% | motif file (matrix) |
pdf |
| 82 |  | Zfp809(Zf)/ES-Zfp809-ChIP-Seq(GSE70799)/Homer | 1e-2 | -5.900e+00 | 0.0129 | 23.0 | 0.77% | 6.6 | 0.25% | motif file (matrix) |
pdf |
| 83 |  | Pho2(bHLH)/Yeast-Pho2-ChIP-Seq(GSE29506)/Homer | 1e-2 | -5.413e+00 | 0.0208 | 34.0 | 1.13% | 13.5 | 0.50% | motif file (matrix) |
pdf |
| 84 |  | SeqBias: CG-repeat | 1e-2 | -5.331e+00 | 0.0223 | 127.0 | 4.23% | 79.0 | 2.92% | motif file (matrix) |
pdf |
| 85 |  | PRDM9(Zf)/Testis-DMC1-ChIP-Seq(GSE35498)/Homer | 1e-2 | -5.237e+00 | 0.0242 | 55.0 | 1.83% | 27.2 | 1.00% | motif file (matrix) |
pdf |
| 86 |  | STAT5(Stat)/mCD4+-Stat5-ChIP-Seq(GSE12346)/Homer | 1e-2 | -5.226e+00 | 0.0242 | 52.0 | 1.73% | 25.5 | 0.94% | motif file (matrix) |
pdf |
| 87 |  | PRDM1(Zf)/Hela-PRDM1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.085e+00 | 0.0275 | 50.0 | 1.67% | 24.2 | 0.90% | motif file (matrix) |
pdf |
| 88 |  | SCL(bHLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-2 | -5.033e+00 | 0.0287 | 750.0 | 25.00% | 599.7 | 22.19% | motif file (matrix) |
pdf |
| 89 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.958e+00 | 0.0306 | 78.0 | 2.60% | 44.9 | 1.66% | motif file (matrix) |
pdf |
| 90 |  | E-box/Drosophila-Promoters/Homer | 1e-2 | -4.947e+00 | 0.0306 | 31.0 | 1.03% | 12.9 | 0.48% | motif file (matrix) |
pdf |
| 91 |  | Rfx5(HTH)/GM12878-Rfx5-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.669e+00 | 0.0399 | 38.0 | 1.27% | 17.5 | 0.65% | motif file (matrix) |
pdf |
| 92 |  | Nr5a2(NR)/mES-Nr5a2-ChIP-Seq(GSE19019)/Homer | 1e-2 | -4.639e+00 | 0.0407 | 57.0 | 1.90% | 30.1 | 1.11% | motif file (matrix) |
pdf |























































































































































































