| p-value: | 1e-7 |
| log p-value: | -1.761e+01 |
| Information Content per bp: | 1.825 |
| Number of Target Sequences with motif | 35.0 |
| Percentage of Target Sequences with motif | 1.17% |
| Number of Background Sequences with motif | 2.6 |
| Percentage of Background Sequences with motif | 0.10% |
| Average Position of motif in Targets | 34.6 +/- 19.7bp |
| Average Position of motif in Background | 20.7 +/- 6.2bp |
| Strand Bias (log2 ratio + to - strand density) | 0.5 |
| Multiplicity (# of sites on avg that occur together) | 1.06 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
SWI5/Literature(Harbison)/Yeast
| Match Rank: | 1 |
| Score: | 0.71
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CCAGCCCTCA
CCAGCA---- |
|


|
|
ACE2/MA0267.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.70
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CCAGCCCTCA
ACCAGCA---- |
|


|
|
TBF1/MA0403.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.68
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CCAGCCCTCA
--AACCCTAA |
|


|
|
ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 4 |
| Score: | 0.68
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CCAGCCCTCA
CNAGGCCT-- |
|


|
|
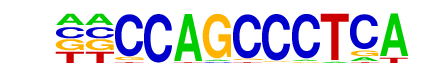
SWI5/MA0402.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.68
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CCAGCCCTCA
AACCAGCA---- |
|

|
|
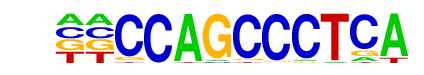
SWI5(MacIsaac)/Yeast
| Match Rank: | 6 |
| Score: | 0.67
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CCAGCCCTCA
AACCAGCA---- |
|

|
|
THAP1/MA0597.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.66
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CCAGCCCTCA
-CTGCCCGCA |
|


|
|
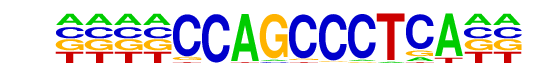
PB0110.1_Bcl6b_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.64
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CCAGCCCTCA--
ATCCCCGCCCCTAAAA |
|

|
|
ZNF711(Zf)/SHSY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer
| Match Rank: | 9 |
| Score: | 0.63
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CCAGCCCTCA
CTAGGCCT-- |
|


|
|
Unknown2/Drosophila-Promoters/Homer
| Match Rank: | 10 |
| Score: | 0.63
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CCAGCCCTCA
-CATCMCTA- |
|


|
|





















