| p-value: | 1e-36 |
| log p-value: | -8.506e+01 |
| Information Content per bp: | 1.785 |
| Number of Target Sequences with motif | 363.0 |
| Percentage of Target Sequences with motif | 12.10% |
| Number of Background Sequences with motif | 89.1 |
| Percentage of Background Sequences with motif | 3.27% |
| Average Position of motif in Targets | 37.0 +/- 19.9bp |
| Average Position of motif in Background | 33.6 +/- 22.8bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.07 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer
| Match Rank: | 1 |
| Score: | 0.83
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TTTCAGTT--
RSTTTCRSTTTC |
|


|
|
Initiator/Drosophila-Promoters/Homer
| Match Rank: | 2 |
| Score: | 0.82
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TTTCAGTT-
-NTCAGTYG |
|


|
|
IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer
| Match Rank: | 3 |
| Score: | 0.82
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TTTCAGTT--
ACTTTCACTTTC |
|


|
|
ISRE(IRF)/ThioMac-LPS-Expression(GSE23622)/Homer
| Match Rank: | 4 |
| Score: | 0.82
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TTTCAGTT--
AGTTTCAGTTTC |
|


|
|
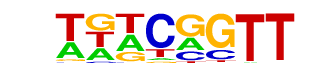
MF0009.1_TRP(MYB)_class/Jaspar
| Match Rank: | 5 |
| Score: | 0.80
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TTTCAGTT
TGTCGGTT |
|

|
|
PU.1:IRF8(ETS:IRF)/pDC-Irf8-ChIP-Seq(GSE66899)/Homer
| Match Rank: | 6 |
| Score: | 0.80
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TTTCAGTT--
ASTTTCACTTCC |
|


|
|
PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 7 |
| Score: | 0.79
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TTTCAGTT---
GTTTCACTTCCG |
|


|
|
blmp-1/MA0537.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.77
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TTTCAGTT--
TTTTCNCTTTT |
|


|
|
IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 9 |
| Score: | 0.77
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TTTCAGTT
TGGTTTCAGT- |
|


|
|
STAT1::STAT2/MA0517.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.76
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----TTTCAGTT---
TCAGTTTCATTTTCC |
|


|
|





















