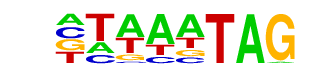
| p-value: | 1e-36 |
| log p-value: | -8.518e+01 |
| Information Content per bp: | 1.621 |
| Number of Target Sequences with motif | 242.0 |
| Percentage of Target Sequences with motif | 28.64% |
| Number of Background Sequences with motif | 35.9 |
| Percentage of Background Sequences with motif | 5.06% |
| Average Position of motif in Targets | 37.6 +/- 20.9bp |
| Average Position of motif in Background | 36.4 +/- 14.8bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.15 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Mef2a(MADS)/HL1-Mef2a.biotin-ChIP-Seq(GSE21529)/Homer
| Match Rank: | 1 |
| Score: | 0.88
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TAAAATAG
CCAAAAATAG |
|


|
|
br(var.2)/MA0011.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.88
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TAAAATAG--
--AAATAGTA |
|


|
|
Mef2c(MADS)/GM12878-Mef2c-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 3 |
| Score: | 0.87
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TAAAATAG-
DCYAAAAATAGM |
|


|
|
Mef2b(MADS)/HEK293-Mef2b.V5-ChIP-Seq(GSE67450)/Homer
| Match Rank: | 4 |
| Score: | 0.85
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TAAAATAG-
KCCAAAAATAGC |
|


|
|
MEF2C/MA0497.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.85
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----TAAAATAG--
ATGCTAAAAATAGAA |
|


|
|
Mef2d(MADS)/Retina-Mef2d-ChIP-Seq(GSE61391)/Homer
| Match Rank: | 6 |
| Score: | 0.82
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TAAAATAG-
GCTAAAAATAGC |
|


|
|
br-Z2/dmmpmm(Bigfoot)/fly
| Match Rank: | 7 |
| Score: | 0.81
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TAAAATAG
-TAAATAG |
|

|
|
br-Z2/dmmpmm(Bergman)/fly
| Match Rank: | 8 |
| Score: | 0.79
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TAAAATAG----
--AAATAGAAAA |
|


|
|
bin/dmmpmm(Bergman)/fly
| Match Rank: | 9 |
| Score: | 0.79
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TAAAATAG
ATAAATA- |
|


|
|
MEF2A/MA0052.3/Jaspar
| Match Rank: | 10 |
| Score: | 0.76
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TAAAATAG-
TCTAAAAATAGA |
|


|
|





















