| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-23 | -5.393e+01 | 0.0000 | 128.0 | 15.15% | 11.0 | 1.56% | motif file (matrix) |
pdf |
| 2 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-22 | -5.276e+01 | 0.0000 | 123.0 | 14.56% | 10.4 | 1.46% | motif file (matrix) |
pdf |
| 3 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-21 | -5.043e+01 | 0.0000 | 116.0 | 13.73% | 9.5 | 1.34% | motif file (matrix) |
pdf |
| 4 |  | Fra1(bZIP)/BT549-Fra1-ChIP-Seq(GSE46166)/Homer | 1e-20 | -4.829e+01 | 0.0000 | 106.0 | 12.54% | 7.3 | 1.02% | motif file (matrix) |
pdf |
| 5 |  | Fosl2(bZIP)/3T3L1-Fosl2-ChIP-Seq(GSE56872)/Homer | 1e-19 | -4.556e+01 | 0.0000 | 87.0 | 10.30% | 3.4 | 0.48% | motif file (matrix) |
pdf |
| 6 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-18 | -4.332e+01 | 0.0000 | 69.0 | 8.17% | 0.6 | 0.08% | motif file (matrix) |
pdf |
| 7 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-17 | -4.134e+01 | 0.0000 | 137.0 | 16.21% | 23.3 | 3.28% | motif file (matrix) |
pdf |
| 8 |  | Mef2b(MADS)/HEK293-Mef2b.V5-ChIP-Seq(GSE67450)/Homer | 1e-17 | -4.116e+01 | 0.0000 | 163.0 | 19.29% | 35.5 | 5.01% | motif file (matrix) |
pdf |
| 9 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-16 | -3.898e+01 | 0.0000 | 123.0 | 14.56% | 19.5 | 2.75% | motif file (matrix) |
pdf |
| 10 |  | Mef2a(MADS)/HL1-Mef2a.biotin-ChIP-Seq(GSE21529)/Homer | 1e-14 | -3.339e+01 | 0.0000 | 105.0 | 12.43% | 16.8 | 2.37% | motif file (matrix) |
pdf |
| 11 |  | Mef2d(MADS)/Retina-Mef2d-ChIP-Seq(GSE61391)/Homer | 1e-14 | -3.291e+01 | 0.0000 | 70.0 | 8.28% | 4.9 | 0.69% | motif file (matrix) |
pdf |
| 12 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-14 | -3.261e+01 | 0.0000 | 113.0 | 13.37% | 20.7 | 2.91% | motif file (matrix) |
pdf |
| 13 |  | Mef2c(MADS)/GM12878-Mef2c-ChIP-Seq(GSE32465)/Homer | 1e-12 | -2.951e+01 | 0.0000 | 122.0 | 14.44% | 27.9 | 3.94% | motif file (matrix) |
pdf |
| 14 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer | 1e-11 | -2.561e+01 | 0.0000 | 103.0 | 12.19% | 22.7 | 3.21% | motif file (matrix) |
pdf |
| 15 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-10 | -2.516e+01 | 0.0000 | 74.0 | 8.76% | 10.9 | 1.54% | motif file (matrix) |
pdf |
| 16 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-10 | -2.362e+01 | 0.0000 | 43.0 | 5.09% | 1.4 | 0.19% | motif file (matrix) |
pdf |
| 17 | 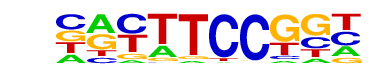 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-9 | -2.195e+01 | 0.0000 | 82.0 | 9.70% | 16.8 | 2.37% | motif file (matrix) |
pdf |
| 18 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-8 | -2.018e+01 | 0.0000 | 97.0 | 11.48% | 25.1 | 3.54% | motif file (matrix) |
pdf |
| 19 |  | IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer | 1e-7 | -1.722e+01 | 0.0000 | 84.0 | 9.94% | 22.2 | 3.13% | motif file (matrix) |
pdf |
| 20 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-7 | -1.709e+01 | 0.0000 | 99.0 | 11.72% | 30.3 | 4.28% | motif file (matrix) |
pdf |
| 21 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-7 | -1.696e+01 | 0.0000 | 42.0 | 4.97% | 4.6 | 0.65% | motif file (matrix) |
pdf |
| 22 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-7 | -1.642e+01 | 0.0000 | 41.0 | 4.85% | 4.7 | 0.67% | motif file (matrix) |
pdf |
| 23 | 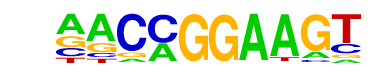 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-7 | -1.642e+01 | 0.0000 | 84.0 | 9.94% | 23.9 | 3.38% | motif file (matrix) |
pdf |
| 24 |  | SpiB(ETS)/OCILY3-SPIB-ChIP-Seq(GSE56857)/Homer | 1e-6 | -1.579e+01 | 0.0000 | 30.0 | 3.55% | 2.0 | 0.28% | motif file (matrix) |
pdf |
| 25 |  | PU.1:IRF8(ETS:IRF)/pDC-Irf8-ChIP-Seq(GSE66899)/Homer | 1e-5 | -1.319e+01 | 0.0000 | 29.0 | 3.43% | 2.5 | 0.36% | motif file (matrix) |
pdf |
| 26 |  | USF1(bHLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-5 | -1.261e+01 | 0.0000 | 31.0 | 3.67% | 4.0 | 0.56% | motif file (matrix) |
pdf |
| 27 |  | CLOCK(bHLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-5 | -1.242e+01 | 0.0001 | 41.0 | 4.85% | 7.4 | 1.05% | motif file (matrix) |
pdf |
| 28 |  | ETS(ETS)/Promoter/Homer | 1e-5 | -1.226e+01 | 0.0001 | 24.0 | 2.84% | 2.0 | 0.28% | motif file (matrix) |
pdf |
| 29 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.194e+01 | 0.0001 | 40.0 | 4.73% | 7.4 | 1.05% | motif file (matrix) |
pdf |
| 30 |  | Max(bHLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.179e+01 | 0.0001 | 42.0 | 4.97% | 8.9 | 1.26% | motif file (matrix) |
pdf |
| 31 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer | 1e-5 | -1.152e+01 | 0.0001 | 26.0 | 3.08% | 2.4 | 0.34% | motif file (matrix) |
pdf |
| 32 | 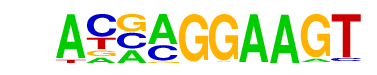 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-4 | -1.149e+01 | 0.0001 | 50.0 | 5.92% | 12.5 | 1.77% | motif file (matrix) |
pdf |
| 33 |  | Initiator/Drosophila-Promoters/Homer | 1e-4 | -1.147e+01 | 0.0001 | 177.0 | 20.95% | 90.8 | 12.81% | motif file (matrix) |
pdf |
| 34 |  | Etv2(ETS)/ES-ER71-ChIP-Seq(GSE59402)/Homer(0.967) | 1e-4 | -1.146e+01 | 0.0001 | 56.0 | 6.63% | 15.0 | 2.12% | motif file (matrix) |
pdf |
| 35 |  | bZIP:IRF(bZIP,IRF)/Th17-BatF-ChIP-Seq(GSE39756)/Homer | 1e-4 | -1.086e+01 | 0.0002 | 64.0 | 7.57% | 20.0 | 2.82% | motif file (matrix) |
pdf |
| 36 |  | BMAL1(bHLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-4 | -1.068e+01 | 0.0002 | 88.0 | 10.41% | 34.6 | 4.89% | motif file (matrix) |
pdf |
| 37 | 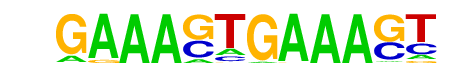 | IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer | 1e-4 | -1.042e+01 | 0.0003 | 24.0 | 2.84% | 3.0 | 0.42% | motif file (matrix) |
pdf |
| 38 |  | FHY3(FAR1)/Arabidopsis-FHY3-ChIP-Seq(GSE30711)/Homer | 1e-4 | -1.042e+01 | 0.0003 | 17.0 | 2.01% | 0.4 | 0.05% | motif file (matrix) |
pdf |
| 39 |  | Cbf1(bHLH)/Yeast-Cbf1-ChIP-Seq(GSE29506)/Homer | 1e-4 | -9.803e+00 | 0.0005 | 16.0 | 1.89% | 1.0 | 0.14% | motif file (matrix) |
pdf |
| 40 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-4 | -9.436e+00 | 0.0008 | 43.0 | 5.09% | 11.9 | 1.68% | motif file (matrix) |
pdf |
| 41 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-4 | -9.348e+00 | 0.0008 | 58.0 | 6.86% | 19.1 | 2.69% | motif file (matrix) |
pdf |
| 42 | 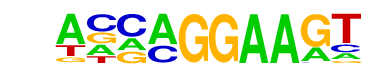 | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-4 | -9.240e+00 | 0.0009 | 70.0 | 8.28% | 26.4 | 3.72% | motif file (matrix) |
pdf |
| 43 |  | Tgif2(Homeobox)/mES-Tgif2-ChIP-Seq(GSE55404)/Homer | 1e-3 | -9.132e+00 | 0.0010 | 163.0 | 19.29% | 87.8 | 12.39% | motif file (matrix) |
pdf |
| 44 | 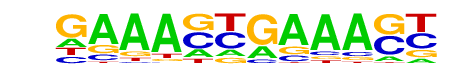 | IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer | 1e-3 | -8.801e+00 | 0.0013 | 21.0 | 2.49% | 2.6 | 0.37% | motif file (matrix) |
pdf |
| 45 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.570e+00 | 0.0016 | 14.0 | 1.66% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 46 |  | c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.439e+00 | 0.0018 | 23.0 | 2.72% | 3.4 | 0.49% | motif file (matrix) |
pdf |
| 47 |  | n-Myc(bHLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-3 | -8.234e+00 | 0.0022 | 40.0 | 4.73% | 11.1 | 1.57% | motif file (matrix) |
pdf |
| 48 |  | Usf2(bHLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-3 | -7.936e+00 | 0.0029 | 22.0 | 2.60% | 3.8 | 0.54% | motif file (matrix) |
pdf |
| 49 |  | E-box/Arabidopsis-Promoters/Homer | 1e-3 | -7.824e+00 | 0.0032 | 37.0 | 4.38% | 10.1 | 1.43% | motif file (matrix) |
pdf |
| 50 |  | AMYB(HTH)/Testes-AMYB-ChIP-Seq(GSE44588)/Homer | 1e-3 | -7.765e+00 | 0.0033 | 94.0 | 11.12% | 44.5 | 6.28% | motif file (matrix) |
pdf |
| 51 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan_et_al.)/Homer | 1e-3 | -7.605e+00 | 0.0038 | 104.0 | 12.31% | 51.1 | 7.21% | motif file (matrix) |
pdf |
| 52 |  | Pho2(bHLH)/Yeast-Pho2-ChIP-Seq(GSE29506)/Homer | 1e-3 | -7.582e+00 | 0.0038 | 26.0 | 3.08% | 5.6 | 0.78% | motif file (matrix) |
pdf |
| 53 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-3 | -7.582e+00 | 0.0038 | 26.0 | 3.08% | 5.6 | 0.79% | motif file (matrix) |
pdf |
| 54 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-3 | -7.497e+00 | 0.0040 | 40.0 | 4.73% | 12.1 | 1.70% | motif file (matrix) |
pdf |
| 55 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-3 | -7.494e+00 | 0.0040 | 28.0 | 3.31% | 6.6 | 0.94% | motif file (matrix) |
pdf |
| 56 |  | BMYB(HTH)/Hela-BMYB-ChIP-Seq(GSE27030)/Homer | 1e-3 | -7.362e+00 | 0.0044 | 97.0 | 11.48% | 47.2 | 6.66% | motif file (matrix) |
pdf |
| 57 |  | SPCH(bHLH)/Seedling-SPCH-ChIP-Seq(GSE57497)/Homer | 1e-3 | -7.145e+00 | 0.0054 | 60.0 | 7.10% | 24.0 | 3.39% | motif file (matrix) |
pdf |
| 58 |  | SOC1(MADS)/Seedling-SOC1-ChIP-Seq(GSE45846)/Homer | 1e-2 | -6.781e+00 | 0.0076 | 22.0 | 2.60% | 4.7 | 0.67% | motif file (matrix) |
pdf |
| 59 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-2 | -6.725e+00 | 0.0079 | 11.0 | 1.30% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 60 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.661e+00 | 0.0083 | 34.0 | 4.02% | 10.4 | 1.46% | motif file (matrix) |
pdf |
| 61 |  | ZNF467(Zf)/HEK293-ZNF467.GFP-ChIP-Seq(GSE58341)/Homer | 1e-2 | -6.629e+00 | 0.0084 | 26.0 | 3.08% | 6.1 | 0.86% | motif file (matrix) |
pdf |
| 62 |  | Klf9(Zf)/GBM-Klf9-ChIP-Seq(GSE62211)/Homer | 1e-2 | -6.563e+00 | 0.0085 | 14.0 | 1.66% | 1.1 | 0.16% | motif file (matrix) |
pdf |
| 63 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.563e+00 | 0.0085 | 14.0 | 1.66% | 1.1 | 0.16% | motif file (matrix) |
pdf |
| 64 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.563e+00 | 0.0085 | 14.0 | 1.66% | 1.7 | 0.24% | motif file (matrix) |
pdf |
| 65 |  | PIF5ox(bHLH)/Arabidopsis-PIF5ox-ChIP-Seq(GSE35062)/Homer | 1e-2 | -6.429e+00 | 0.0096 | 51.0 | 6.04% | 20.6 | 2.90% | motif file (matrix) |
pdf |
| 66 |  | PRDM1(Zf)/Hela-PRDM1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.285e+00 | 0.0109 | 33.0 | 3.91% | 10.7 | 1.51% | motif file (matrix) |
pdf |
| 67 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-2 | -6.212e+00 | 0.0116 | 29.0 | 3.43% | 8.7 | 1.23% | motif file (matrix) |
pdf |
| 68 |  | HLH-1(bHLH)/cElegans-Embryo-HLH1-ChIP-Seq(modEncode)/Homer | 1e-2 | -6.198e+00 | 0.0116 | 27.0 | 3.20% | 7.2 | 1.02% | motif file (matrix) |
pdf |
| 69 |  | PIF4(bHLH)/Seedling-PIF4-ChIP-Seq(GSE35315)/Homer | 1e-2 | -6.078e+00 | 0.0129 | 58.0 | 6.86% | 25.3 | 3.57% | motif file (matrix) |
pdf |
| 70 |  | SeqBias: CA-repeat | 1e-2 | -6.004e+00 | 0.0136 | 272.0 | 32.19% | 181.8 | 25.65% | motif file (matrix) |
pdf |
| 71 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-2 | -5.984e+00 | 0.0137 | 64.0 | 7.57% | 29.8 | 4.21% | motif file (matrix) |
pdf |
| 72 |  | MyoD(bHLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-2 | -5.977e+00 | 0.0137 | 34.0 | 4.02% | 11.7 | 1.64% | motif file (matrix) |
pdf |
| 73 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-2 | -5.951e+00 | 0.0138 | 41.0 | 4.85% | 15.7 | 2.22% | motif file (matrix) |
pdf |
| 74 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-2 | -5.680e+00 | 0.0178 | 15.0 | 1.78% | 2.4 | 0.34% | motif file (matrix) |
pdf |
| 75 |  | NFkB-p50,p52(RHD)/Monocyte-p50-ChIP-Chip(Schreiber_et_al.)/Homer | 1e-2 | -5.497e+00 | 0.0212 | 9.0 | 1.07% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 76 |  | NFAT:AP1(RHD,bZIP)/Jurkat-NFATC1-ChIP-Seq(Jolma_et_al.)/Homer | 1e-2 | -5.465e+00 | 0.0215 | 12.0 | 1.42% | 1.4 | 0.20% | motif file (matrix) |
pdf |
| 77 |  | SeqBias: CG-repeat | 1e-2 | -5.452e+00 | 0.0216 | 109.0 | 12.90% | 61.2 | 8.64% | motif file (matrix) |
pdf |
| 78 |  | Meis1(Homeobox)/MastCells-Meis1-ChIP-Seq(GSE48085)/Homer | 1e-2 | -5.298e+00 | 0.0248 | 69.0 | 8.17% | 34.1 | 4.81% | motif file (matrix) |
pdf |
| 79 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-2 | -5.288e+00 | 0.0248 | 111.0 | 13.14% | 63.8 | 9.00% | motif file (matrix) |
pdf |
| 80 |  | ISRE(IRF)/ThioMac-LPS-Expression(GSE23622)/Homer | 1e-2 | -5.181e+00 | 0.0272 | 14.0 | 1.66% | 2.3 | 0.32% | motif file (matrix) |
pdf |
| 81 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-2 | -5.179e+00 | 0.0272 | 50.0 | 5.92% | 22.3 | 3.14% | motif file (matrix) |
pdf |
| 82 |  | IBL1(bHLH)/Seedling-IBL1-ChIP-Seq(GSE51120)/Homer | 1e-2 | -5.156e+00 | 0.0272 | 89.0 | 10.53% | 48.4 | 6.83% | motif file (matrix) |
pdf |
| 83 |  | IRF:BATF(IRF:bZIP)/pDC-Irf8-ChIP-Seq(GSE66899)/Homer | 1e-2 | -5.129e+00 | 0.0276 | 28.0 | 3.31% | 9.7 | 1.37% | motif file (matrix) |
pdf |
| 84 |  | SCL(bHLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-2 | -5.057e+00 | 0.0293 | 175.0 | 20.71% | 111.8 | 15.77% | motif file (matrix) |
pdf |
| 85 |  | ZNF711(Zf)/SHSY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer | 1e-2 | -5.020e+00 | 0.0297 | 51.0 | 6.04% | 23.1 | 3.26% | motif file (matrix) |
pdf |
| 86 |  | FoxL2(Forkhead)/Ovary-FoxL2-ChIP-Seq(GSE60858)/Homer | 1e-2 | -5.020e+00 | 0.0297 | 51.0 | 6.04% | 23.4 | 3.30% | motif file (matrix) |
pdf |
| 87 |  | Pho4(bHLH)/Yeast-Pho4-ChIP-Seq(GSE29506)/Homer | 1e-2 | -4.982e+00 | 0.0305 | 18.0 | 2.13% | 4.8 | 0.68% | motif file (matrix) |
pdf |
| 88 |  | KLF10(Zf)/HEK293-KLF10.GFP-ChIP-Seq(GSE58341)/Homer | 1e-2 | -4.924e+00 | 0.0320 | 11.0 | 1.30% | 1.6 | 0.22% | motif file (matrix) |
pdf |
| 89 |  | E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer | 1e-2 | -4.884e+00 | 0.0320 | 8.0 | 0.95% | 0.4 | 0.05% | motif file (matrix) |
pdf |
| 90 |  | ETS:RUNX(ETS,Runt)/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -4.884e+00 | 0.0320 | 8.0 | 0.95% | 0.3 | 0.04% | motif file (matrix) |
pdf |
| 91 |  | CEBP:CEBP(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-2 | -4.884e+00 | 0.0320 | 8.0 | 0.95% | 0.5 | 0.08% | motif file (matrix) |
pdf |
| 92 |  | Unknown4/Drosophila-Promoters/Homer | 1e-2 | -4.884e+00 | 0.0320 | 8.0 | 0.95% | 0.8 | 0.11% | motif file (matrix) |
pdf |
| 93 | 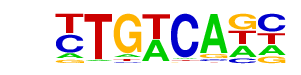 | Tgif1(Homeobox)/mES-Tgif1-ChIP-Seq(GSE55404)/Homer | 1e-2 | -4.852e+00 | 0.0325 | 143.0 | 16.92% | 88.1 | 12.43% | motif file (matrix) |
pdf |
| 94 |  | c-Myc(bHLH)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-2 | -4.702e+00 | 0.0374 | 25.0 | 2.96% | 8.3 | 1.18% | motif file (matrix) |
pdf |
| 95 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-2 | -4.689e+00 | 0.0375 | 13.0 | 1.54% | 2.0 | 0.28% | motif file (matrix) |
pdf |
| 96 |  | CHR(?)/Hela-CellCycle-Expression/Homer | 1e-2 | -4.616e+00 | 0.0399 | 40.0 | 4.73% | 18.0 | 2.53% | motif file (matrix) |
pdf |































































































































































































