





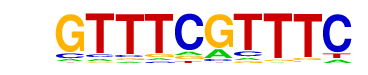




| p-value: | 1e-94 |
| log p-value: | -2.187e+02 |
| Information Content per bp: | 1.490 |
| Number of Target Sequences with motif | 1470.0 |
| Percentage of Target Sequences with motif | 49.00% |
| Number of Background Sequences with motif | 573.2 |
| Percentage of Background Sequences with motif | 22.42% |
| Average Position of motif in Targets | 36.1 +/- 20.0bp |
| Average Position of motif in Background | 37.5 +/- 24.6bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.32 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.644 |  | 1e-69 | -159.288967 | 31.27% | 11.93% | motif file (matrix) |
| 2 | 0.792 |  | 1e-58 | -134.750349 | 16.63% | 3.79% | motif file (matrix) |
| 3 | 0.796 |  | 1e-52 | -121.599014 | 21.97% | 7.51% | motif file (matrix) |
| 4 | 0.728 |  | 1e-45 | -105.839173 | 12.93% | 2.84% | motif file (matrix) |
| 5 | 0.697 |  | 1e-24 | -57.066513 | 11.80% | 4.27% | motif file (matrix) |
| 6 | 0.640 |  | 1e-22 | -51.257733 | 8.43% | 2.52% | motif file (matrix) |
| 7 | 0.605 |  | 1e-18 | -41.905542 | 4.03% | 0.60% | motif file (matrix) |
| 8 | 0.627 |  | 1e-14 | -32.964926 | 2.23% | 0.13% | motif file (matrix) |
| 9 | 0.643 |  | 1e-14 | -32.580550 | 3.33% | 0.58% | motif file (matrix) |