





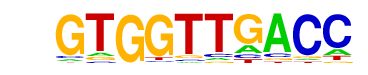
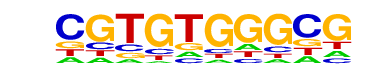


| p-value: | 1e-94 |
| log p-value: | -2.184e+02 |
| Information Content per bp: | 1.740 |
| Number of Target Sequences with motif | 1016.0 |
| Percentage of Target Sequences with motif | 33.87% |
| Number of Background Sequences with motif | 279.0 |
| Percentage of Background Sequences with motif | 10.92% |
| Average Position of motif in Targets | 36.4 +/- 20.5bp |
| Average Position of motif in Background | 36.7 +/- 27.5bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.16 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.926 |  | 1e-75 | -174.117272 | 17.70% | 3.08% | motif file (matrix) |
| 2 | 0.959 |  | 1e-70 | -161.982264 | 15.90% | 2.55% | motif file (matrix) |
| 3 | 0.630 |  | 1e-23 | -54.646107 | 8.00% | 2.12% | motif file (matrix) |
| 4 | 0.649 |  | 1e-18 | -41.628268 | 3.10% | 0.25% | motif file (matrix) |
| 5 | 0.656 |  | 1e-13 | -30.367104 | 2.53% | 0.29% | motif file (matrix) |
| 6 | 0.639 |  | 1e-11 | -27.172608 | 1.90% | 0.15% | motif file (matrix) |
| 7 | 0.757 |  | 1e-11 | -26.082508 | 2.27% | 0.31% | motif file (matrix) |
| 8 | 0.638 |  | 1e-7 | -16.127102 | 1.87% | 0.41% | motif file (matrix) |