| p-value: | 1e-18 |
| log p-value: | -4.233e+01 |
| Information Content per bp: | 1.656 |
| Number of Target Sequences with motif | 122.0 |
| Percentage of Target Sequences with motif | 4.07% |
| Number of Background Sequences with motif | 11.5 |
| Percentage of Background Sequences with motif | 0.51% |
| Average Position of motif in Targets | 32.0 +/- 22.5bp |
| Average Position of motif in Background | 43.9 +/- 11.0bp |
| Strand Bias (log2 ratio + to - strand density) | -0.8 |
| Multiplicity (# of sites on avg that occur together) | 1.02 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
INO4(MacIsaac)/Yeast
| Match Rank: | 1 |
| Score: | 0.79
| | Offset: | 0
| | Orientation: | reverse strand |
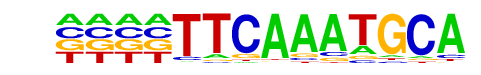
| Alignment: | TTCAAATGCA
TTCACATGC- |
|


|
|
INO2(MacIsaac)/Yeast
| Match Rank: | 2 |
| Score: | 0.79
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TTCAAATGCA
TTCACATGC- |
|


|
|
INO4/MA0322.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.78
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TTCAAATGCA
TTCACATGC- |
|


|
|
INO2/MA0321.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.77
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TTCAAATGCA
TTCACATGC- |
|


|
|
CST6(MacIsaac)/Yeast
| Match Rank: | 5 |
| Score: | 0.75
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TTCAAATGCA
--NAAATGCA |
|


|
|
LIN54/MA0619.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.73
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TTCAAATGCA
NATTCAAAT--- |
|


|
|
CHR(?)/Hela-CellCycle-Expression/Homer
| Match Rank: | 7 |
| Score: | 0.72
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----TTCAAATGCA
CGGTTTCAAA---- |
|

|
|
INO4/INO4_YPD/4-INO4,37-INO2(Harbison)/Yeast
| Match Rank: | 8 |
| Score: | 0.70
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TTCAAATGCA
TTTTCACATG-- |
|


|
|
RME1(MacIsaac)/Yeast
| Match Rank: | 9 |
| Score: | 0.70
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TTCAAATGCA
TCCAAAGGAA |
|


|
|
RME1/MA0370.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.69
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TTCAAATGCA
TCCAAAGGAA |
|


|
|





















