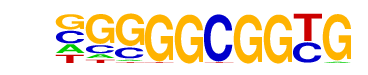| p-value: | 1e-29 |
| log p-value: | -6.865e+01 |
| Information Content per bp: | 1.823 |
| Number of Target Sequences with motif | 130.0 |
| Percentage of Target Sequences with motif | 11.56% |
| Number of Background Sequences with motif | 1.0 |
| Percentage of Background Sequences with motif | 0.13% |
| Average Position of motif in Targets | 41.0 +/- 18.8bp |
| Average Position of motif in Background | 0.0 +/- 0.0bp |
| Strand Bias (log2 ratio + to - strand density) | -0.7 |
| Multiplicity (# of sites on avg that occur together) | 1.04 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer
| Match Rank: | 1 |
| Score: | 0.81
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CGGCGGTT-
-GGCVGTTR |
|


|
|
Run/dmmpmm(Papatsenko)/fly
| Match Rank: | 2 |
| Score: | 0.80
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CGGCGGTT
-GGCGGTG |
|


|
|
Gam1/MA0034.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.80
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CGGCGGTT---
-GGCGGTTGTC |
|


|
|
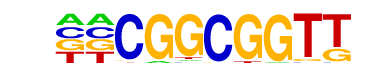
ERF4/MA0992.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.78
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CGGCGGTT
GGCGGCGG-- |
|

|
|
GAMYB(MYB)/Hordeum vulgare/AthaMap
| Match Rank: | 5 |
| Score: | 0.77
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CGGCGGTT-
-GGCGGTTG |
|


|
|
ERF069/MA0997.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.76
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CGGCGGTT
NGGCGGCGN-- |
|


|
|
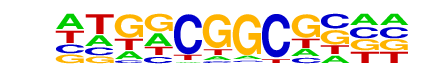
ERF11/MA1001.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.76
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----CGGCGGTT
NTGNCGGCGN-- |
|

|
|
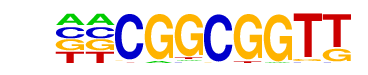
ABI4(1)(AP2/EREBP)/Zea mays/AthaMap
| Match Rank: | 8 |
| Score: | 0.76
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CGGCGGTT
NGGGGCGGTG |
|
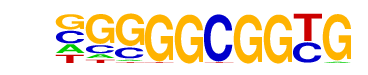
|
|
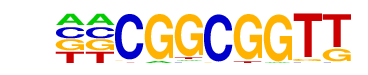
IME1(MacIsaac)/Yeast
| Match Rank: | 9 |
| Score: | 0.76
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CGGCGGTT
CTCGGCGG-- |
|

|
|
UME6(MacIsaac)/Yeast
| Match Rank: | 10 |
| Score: | 0.75
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CGGCGGTT-
TCGGCGGCTA |
|


|
|