| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Sox10(HMG)/SciaticNerve-Sox3-ChIP-Seq(GSE35132)/Homer | 1e-27 | -6.270e+01 | 0.0000 | 202.0 | 17.96% | 19.7 | 2.67% | motif file (matrix) |
pdf |
| 2 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-24 | -5.660e+01 | 0.0000 | 217.0 | 19.29% | 28.3 | 3.84% | motif file (matrix) |
pdf |
| 3 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-22 | -5.268e+01 | 0.0000 | 181.0 | 16.09% | 19.2 | 2.60% | motif file (matrix) |
pdf |
| 4 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-18 | -4.147e+01 | 0.0000 | 119.0 | 10.58% | 8.4 | 1.14% | motif file (matrix) |
pdf |
| 5 |  | Sox4(HMG)/proB-Sox4-ChIP-Seq(GSE50066)/Homer | 1e-16 | -3.779e+01 | 0.0000 | 115.0 | 10.22% | 9.8 | 1.32% | motif file (matrix) |
pdf |
| 6 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-13 | -3.054e+01 | 0.0000 | 71.0 | 6.31% | 2.7 | 0.36% | motif file (matrix) |
pdf |
| 7 |  | Sox15(HMG)/CPA-Sox15-ChIP-Seq(GSE62909)/Homer | 1e-12 | -2.812e+01 | 0.0000 | 110.0 | 9.78% | 14.2 | 1.92% | motif file (matrix) |
pdf |
| 8 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-12 | -2.808e+01 | 0.0000 | 83.0 | 7.38% | 6.4 | 0.87% | motif file (matrix) |
pdf |
| 9 |  | Egr1(Zf)/K562-Egr1-ChIP-Seq(GSE32465)/Homer | 1e-11 | -2.626e+01 | 0.0000 | 67.0 | 5.96% | 4.0 | 0.54% | motif file (matrix) |
pdf |
| 10 |  | IBL1(bHLH)/Seedling-IBL1-ChIP-Seq(GSE51120)/Homer | 1e-11 | -2.617e+01 | 0.0000 | 117.0 | 10.40% | 18.4 | 2.49% | motif file (matrix) |
pdf |
| 11 |  | SCL(bHLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-10 | -2.482e+01 | 0.0000 | 305.0 | 27.11% | 105.8 | 14.34% | motif file (matrix) |
pdf |
| 12 |  | TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer | 1e-10 | -2.395e+01 | 0.0000 | 81.0 | 7.20% | 8.1 | 1.10% | motif file (matrix) |
pdf |
| 13 |  | E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer | 1e-9 | -2.147e+01 | 0.0000 | 42.0 | 3.73% | 0.8 | 0.11% | motif file (matrix) |
pdf |
| 14 |  | TEAD2(TEA)/Py2T-Tead2-ChIP-Seq(GSE55709)/Homer | 1e-8 | -1.874e+01 | 0.0000 | 47.0 | 4.18% | 2.7 | 0.37% | motif file (matrix) |
pdf |
| 15 |  | Unknown-ESC-element(?)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-7 | -1.839e+01 | 0.0000 | 68.0 | 6.04% | 8.4 | 1.14% | motif file (matrix) |
pdf |
| 16 |  | SeqBias: polyA-repeat | 1e-7 | -1.765e+01 | 0.0000 | 805.0 | 71.56% | 436.9 | 59.25% | motif file (matrix) |
pdf |
| 17 |  | Zic(Zf)/Cerebellum-ZIC1.2-ChIP-Seq(GSE60731)/Homer | 1e-7 | -1.726e+01 | 0.0000 | 80.0 | 7.11% | 13.9 | 1.88% | motif file (matrix) |
pdf |
| 18 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-7 | -1.683e+01 | 0.0000 | 43.0 | 3.82% | 2.1 | 0.28% | motif file (matrix) |
pdf |
| 19 |  | Sox9(HMG)/Limb-SOX9-ChIP-Seq(GSE73225)/Homer | 1e-7 | -1.633e+01 | 0.0000 | 69.0 | 6.13% | 10.5 | 1.43% | motif file (matrix) |
pdf |
| 20 |  | Oct4(POU,Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-6 | -1.543e+01 | 0.0000 | 44.0 | 3.91% | 3.8 | 0.51% | motif file (matrix) |
pdf |
| 21 |  | TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer | 1e-6 | -1.481e+01 | 0.0000 | 53.0 | 4.71% | 6.3 | 0.85% | motif file (matrix) |
pdf |
| 22 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan_et_al.)/Homer | 1e-6 | -1.455e+01 | 0.0000 | 98.0 | 8.71% | 23.7 | 3.21% | motif file (matrix) |
pdf |
| 23 |  | ZNF467(Zf)/HEK293-ZNF467.GFP-ChIP-Seq(GSE58341)/Homer | 1e-6 | -1.450e+01 | 0.0000 | 93.0 | 8.27% | 21.4 | 2.90% | motif file (matrix) |
pdf |
| 24 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-5 | -1.268e+01 | 0.0001 | 99.0 | 8.80% | 27.0 | 3.66% | motif file (matrix) |
pdf |
| 25 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-5 | -1.262e+01 | 0.0001 | 34.0 | 3.02% | 2.1 | 0.28% | motif file (matrix) |
pdf |
| 26 |  | OCT4-SOX2-TCF-NANOG(POU,Homeobox,HMG)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-5 | -1.219e+01 | 0.0001 | 24.0 | 2.13% | 0.5 | 0.07% | motif file (matrix) |
pdf |
| 27 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-5 | -1.187e+01 | 0.0001 | 139.0 | 12.36% | 46.9 | 6.36% | motif file (matrix) |
pdf |
| 28 |  | Trl(Zf)/S2-GAGAfactor-ChIP-Seq(GSE40646)/Homer | 1e-5 | -1.164e+01 | 0.0001 | 168.0 | 14.93% | 61.6 | 8.35% | motif file (matrix) |
pdf |
| 29 |  | Etv2(ETS)/ES-ER71-ChIP-Seq(GSE59402)/Homer(0.967) | 1e-5 | -1.156e+01 | 0.0001 | 48.0 | 4.27% | 7.7 | 1.04% | motif file (matrix) |
pdf |
| 30 |  | Egr2(Zf)/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-4 | -1.117e+01 | 0.0002 | 22.0 | 1.96% | 0.2 | 0.03% | motif file (matrix) |
pdf |
| 31 |  | KLF10(Zf)/HEK293-KLF10.GFP-ChIP-Seq(GSE58341)/Homer | 1e-4 | -1.097e+01 | 0.0002 | 34.0 | 3.02% | 3.3 | 0.45% | motif file (matrix) |
pdf |
| 32 |  | Brn1(POU,Homeobox)/NPC-Brn1-ChIP-Seq(GSE35496)/Homer | 1e-4 | -1.097e+01 | 0.0002 | 34.0 | 3.02% | 3.8 | 0.52% | motif file (matrix) |
pdf |
| 33 |  | Oct6(POU,Homeobox)/NPC-Oct6-ChIP-Seq(GSE35496)/Homer | 1e-4 | -1.041e+01 | 0.0004 | 36.0 | 3.20% | 4.7 | 0.63% | motif file (matrix) |
pdf |
| 34 |  | FHY3(FAR1)/Arabidopsis-FHY3-ChIP-Seq(GSE30711)/Homer | 1e-4 | -1.034e+01 | 0.0004 | 29.0 | 2.58% | 2.5 | 0.34% | motif file (matrix) |
pdf |
| 35 |  | SeqBias: CG bias | 1e-4 | -9.680e+00 | 0.0007 | 919.0 | 81.69% | 546.1 | 74.06% | motif file (matrix) |
pdf |
| 36 |  | MyoG(bHLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-3 | -8.184e+00 | 0.0030 | 54.0 | 4.80% | 13.5 | 1.84% | motif file (matrix) |
pdf |
| 37 |  | E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer | 1e-3 | -8.104e+00 | 0.0032 | 16.0 | 1.42% | 0.2 | 0.03% | motif file (matrix) |
pdf |
| 38 |  | HIF-1b(HLH)/T47D-HIF1b-ChIP-Seq(GSE59937)/Homer | 1e-3 | -7.837e+00 | 0.0040 | 38.0 | 3.38% | 7.0 | 0.95% | motif file (matrix) |
pdf |
| 39 |  | Ap4(bHLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-3 | -7.819e+00 | 0.0040 | 73.0 | 6.49% | 22.1 | 3.00% | motif file (matrix) |
pdf |
| 40 |  | Ptf1a(bHLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer | 1e-3 | -7.765e+00 | 0.0041 | 131.0 | 11.64% | 51.6 | 6.99% | motif file (matrix) |
pdf |
| 41 |  | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer | 1e-3 | -7.607e+00 | 0.0047 | 107.0 | 9.51% | 39.7 | 5.38% | motif file (matrix) |
pdf |
| 42 |  | ZNF189(Zf)/HEK293-ZNF189.GFP-ChIP-Seq(GSE58341)/Homer | 1e-3 | -7.569e+00 | 0.0048 | 52.0 | 4.62% | 13.9 | 1.89% | motif file (matrix) |
pdf |
| 43 |  | Pho2(bHLH)/Yeast-Pho2-ChIP-Seq(GSE29506)/Homer | 1e-3 | -7.483e+00 | 0.0051 | 19.0 | 1.69% | 1.7 | 0.22% | motif file (matrix) |
pdf |
| 44 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-3 | -7.366e+00 | 0.0056 | 96.0 | 8.53% | 34.2 | 4.63% | motif file (matrix) |
pdf |
| 45 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.251e+00 | 0.0061 | 60.0 | 5.33% | 17.9 | 2.42% | motif file (matrix) |
pdf |
| 46 |  | Lhx3(Homeobox)/Neuron-Lhx3-ChIP-Seq(GSE31456)/Homer | 1e-3 | -7.141e+00 | 0.0067 | 97.0 | 8.62% | 35.9 | 4.86% | motif file (matrix) |
pdf |
| 47 |  | MyoD(bHLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-3 | -7.063e+00 | 0.0070 | 48.0 | 4.27% | 12.0 | 1.63% | motif file (matrix) |
pdf |
| 48 |  | Oct2(POU,Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-2 | -6.770e+00 | 0.0093 | 24.0 | 2.13% | 3.1 | 0.43% | motif file (matrix) |
pdf |
| 49 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-2 | -6.720e+00 | 0.0095 | 75.0 | 6.67% | 25.4 | 3.45% | motif file (matrix) |
pdf |
| 50 |  | Ascl1(bHLH)/NeuralTubes-Ascl1-ChIP-Seq(GSE55840)/Homer | 1e-2 | -6.682e+00 | 0.0097 | 85.0 | 7.56% | 30.3 | 4.11% | motif file (matrix) |
pdf |
| 51 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-2 | -6.578e+00 | 0.0106 | 13.0 | 1.16% | 0.3 | 0.04% | motif file (matrix) |
pdf |
| 52 | 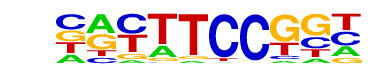 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-2 | -6.553e+00 | 0.0106 | 66.0 | 5.87% | 21.3 | 2.89% | motif file (matrix) |
pdf |
| 53 |  | Lhx2(Homeobox)/HFSC-Lhx2-ChIP-Seq(GSE48068)/Homer | 1e-2 | -6.553e+00 | 0.0106 | 66.0 | 5.87% | 21.0 | 2.85% | motif file (matrix) |
pdf |
| 54 | 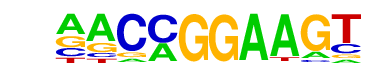 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-2 | -6.469e+00 | 0.0111 | 76.0 | 6.76% | 26.8 | 3.63% | motif file (matrix) |
pdf |
| 55 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-2 | -6.457e+00 | 0.0111 | 80.0 | 7.11% | 28.6 | 3.88% | motif file (matrix) |
pdf |
| 56 |  | Tcf21(bHLH)/ArterySmoothMuscle-Tcf21-ChIP-Seq(GSE61369)/Homer | 1e-2 | -6.431e+00 | 0.0111 | 57.0 | 5.07% | 17.6 | 2.39% | motif file (matrix) |
pdf |
| 57 |  | ZNF165(Zf)/WHIM12-ZNF165-ChIP-Seq(GSE65937)/Homer | 1e-2 | -6.070e+00 | 0.0157 | 12.0 | 1.07% | 0.3 | 0.04% | motif file (matrix) |
pdf |
| 58 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-2 | -5.677e+00 | 0.0228 | 52.0 | 4.62% | 16.9 | 2.30% | motif file (matrix) |
pdf |
| 59 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -5.619e+00 | 0.0238 | 56.0 | 4.98% | 18.4 | 2.49% | motif file (matrix) |
pdf |
| 60 |  | Tcf12(bHLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.598e+00 | 0.0239 | 58.0 | 5.16% | 19.7 | 2.67% | motif file (matrix) |
pdf |
| 61 | 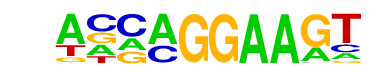 | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-2 | -5.568e+00 | 0.0242 | 62.0 | 5.51% | 21.5 | 2.91% | motif file (matrix) |
pdf |
| 62 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-2 | -5.418e+00 | 0.0277 | 51.0 | 4.53% | 16.1 | 2.19% | motif file (matrix) |
pdf |
| 63 |  | Lhx1(Homeobox)/EmbryoCarcinoma-Lhx1-ChIP-Seq(GSE70957)/Homer | 1e-2 | -5.323e+00 | 0.0300 | 65.0 | 5.78% | 23.0 | 3.12% | motif file (matrix) |
pdf |
| 64 |  | PAX5(Paired,Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.306e+00 | 0.0300 | 23.0 | 2.04% | 4.2 | 0.58% | motif file (matrix) |
pdf |
| 65 |  | MITF(bHLH)/MastCells-MITF-ChIP-Seq(GSE48085)/Homer | 1e-2 | -5.274e+00 | 0.0305 | 28.0 | 2.49% | 6.7 | 0.91% | motif file (matrix) |
pdf |
| 66 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-2 | -5.239e+00 | 0.0311 | 35.0 | 3.11% | 9.5 | 1.29% | motif file (matrix) |
pdf |
| 67 |  | NFkB-p50,p52(RHD)/Monocyte-p50-ChIP-Chip(Schreiber_et_al.)/Homer | 1e-2 | -5.202e+00 | 0.0313 | 14.0 | 1.24% | 1.8 | 0.24% | motif file (matrix) |
pdf |
| 68 |  | NRF(NRF)/Promoter/Homer | 1e-2 | -5.202e+00 | 0.0313 | 14.0 | 1.24% | 1.3 | 0.18% | motif file (matrix) |
pdf |
| 69 |  | E-box/Arabidopsis-Promoters/Homer | 1e-2 | -5.142e+00 | 0.0328 | 17.0 | 1.51% | 2.0 | 0.28% | motif file (matrix) |
pdf |
| 70 |  | FoxL2(Forkhead)/Ovary-FoxL2-ChIP-Seq(GSE60858)/Homer | 1e-2 | -4.944e+00 | 0.0394 | 22.0 | 1.96% | 4.7 | 0.64% | motif file (matrix) |
pdf |
| 71 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-2 | -4.943e+00 | 0.0394 | 34.0 | 3.02% | 9.7 | 1.31% | motif file (matrix) |
pdf |
| 72 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-2 | -4.760e+00 | 0.0461 | 24.0 | 2.13% | 5.7 | 0.77% | motif file (matrix) |
pdf |
| 73 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.757e+00 | 0.0461 | 239.0 | 21.24% | 123.5 | 16.74% | motif file (matrix) |
pdf |
| 74 |  | EKLF(Zf)/Erythrocyte-Klf1-ChIP-Seq(GSE20478)/Homer | 1e-2 | -4.756e+00 | 0.0461 | 13.0 | 1.16% | 1.8 | 0.24% | motif file (matrix) |
pdf |
| 75 |  | HIF2a(bHLH)/785_O-HIF2a-ChIP-Seq(GSE34871)/Homer | 1e-2 | -4.756e+00 | 0.0461 | 13.0 | 1.16% | 1.0 | 0.14% | motif file (matrix) |
pdf |
| 76 |  | Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer | 1e-2 | -4.644e+00 | 0.0490 | 54.0 | 4.80% | 19.9 | 2.69% | motif file (matrix) |
pdf |
| 77 |  | KLF14(Zf)/HEK293-KLF14.GFP-ChIP-Seq(GSE58341)/Homer | 1e-2 | -4.636e+00 | 0.0490 | 228.0 | 20.27% | 117.1 | 15.88% | motif file (matrix) |
pdf |

























































































































































