




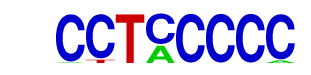


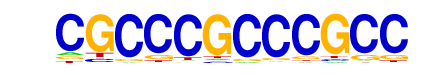
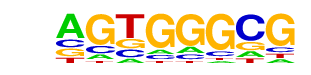




| p-value: | 1e-35 |
| log p-value: | -8.187e+01 |
| Information Content per bp: | 1.723 |
| Number of Target Sequences with motif | 179.0 |
| Percentage of Target Sequences with motif | 5.97% |
| Number of Background Sequences with motif | 5.1 |
| Percentage of Background Sequences with motif | 0.24% |
| Average Position of motif in Targets | 37.6 +/- 19.4bp |
| Average Position of motif in Background | 37.5 +/- 22.5bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.13 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.666 |  | 1e-33 | -77.857466 | 6.60% | 0.54% | motif file (matrix) |
| 2 | 0.832 |  | 1e-31 | -72.784038 | 6.40% | 0.59% | motif file (matrix) |
| 3 | 0.647 |  | 1e-28 | -66.357276 | 10.17% | 2.51% | motif file (matrix) |
| 4 | 0.628 |  | 1e-27 | -63.733632 | 4.50% | 0.15% | motif file (matrix) |
| 5 | 0.637 |  | 1e-25 | -58.500741 | 5.57% | 0.62% | motif file (matrix) |
| 6 | 0.740 |  | 1e-24 | -57.433126 | 7.10% | 1.32% | motif file (matrix) |
| 7 | 0.751 |  | 1e-24 | -56.587268 | 4.67% | 0.33% | motif file (matrix) |
| 8 | 0.609 |  | 1e-23 | -54.809936 | 3.77% | 0.11% | motif file (matrix) |
| 9 | 0.672 |  | 1e-15 | -35.392358 | 2.17% | 0.00% | motif file (matrix) |
| 10 | 0.630 |  | 1e-14 | -33.931500 | 7.33% | 2.52% | motif file (matrix) |
| 11 | 0.668 |  | 1e-12 | -29.736202 | 3.90% | 0.82% | motif file (matrix) |
| 12 | 0.678 |  | 1e-11 | -26.739481 | 2.87% | 0.45% | motif file (matrix) |