| p-value: | 1e-5 |
| log p-value: | -1.370e+01 |
| Information Content per bp: | 1.690 |
| Number of Target Sequences with motif | 92.0 |
| Percentage of Target Sequences with motif | 3.84% |
| Number of Background Sequences with motif | 41.6 |
| Percentage of Background Sequences with motif | 1.65% |
| Average Position of motif in Targets | 39.9 +/- 19.5bp |
| Average Position of motif in Background | 37.6 +/- 20.2bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.14 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
NR4A2/MA0160.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.75
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AGRTCMCG
AAGGTCAC- |
|


|
|
RTG3/Literature(Harbison)/Yeast
| Match Rank: | 2 |
| Score: | 0.73
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AGRTCMCG
-GGTCAC- |
|


|
|
Mitf/MA0620.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.70
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AGRTCMCG---
-GGTCACGTGG |
|


|
|
ZmHOX2a(2)(HD-HOX)/Zea mays/AthaMap
| Match Rank: | 4 |
| Score: | 0.70
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AGRTCMCG
CAGATCA-- |
|


|
|
POL011.1_XCPE1/Jaspar
| Match Rank: | 5 |
| Score: | 0.68
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | AGRTCMCG---
-GGTCCCGCCC |
|


|
|
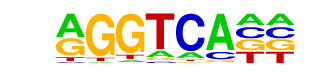
MF0004.1_Nuclear_Receptor_class/Jaspar
| Match Rank: | 6 |
| Score: | 0.67
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGRTCMCG
AGGTCA-- |
|

|
|
Arntl/MA0603.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.67
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AGRTCMCG---
-GGTCACGTGC |
|


|
|
PB0053.1_Rara_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.67
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------AGRTCMCG--
TCTCAAAGGTCACCTG |
|


|
|
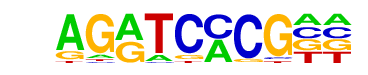
TYE7(MacIsaac)/Yeast
| Match Rank: | 9 |
| Score: | 0.67
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | AGRTCMCG--
--GTCACGTG |
|

|
|
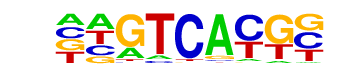
Pax2/MA0067.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.66
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AGRTCMCG-
-AGTCACGC |
|

|
|





















