| p-value: | 1e-10 |
| log p-value: | -2.314e+01 |
| Information Content per bp: | 1.650 |
| Number of Target Sequences with motif | 185.0 |
| Percentage of Target Sequences with motif | 7.71% |
| Number of Background Sequences with motif | 89.3 |
| Percentage of Background Sequences with motif | 3.54% |
| Average Position of motif in Targets | 38.9 +/- 18.4bp |
| Average Position of motif in Background | 35.8 +/- 18.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.05 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
ZNF416(Zf)/HEK293-ZNF416.GFP-ChIP-Seq(GSE58341)/Homer
| Match Rank: | 1 |
| Score: | 0.67
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TTGYCCAGGCTG
-TGCCCAGNHW- |
|


|
|
Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer
| Match Rank: | 2 |
| Score: | 0.63
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TTGYCCAGGCTG
HTTTCCCASG--- |
|


|
|
NFATC2/MA0152.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.61
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TTGYCCAGGCTG
TTTTCCA----- |
|


|
|
nub/dmmpmm(Pollard)/fly
| Match Rank: | 4 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TTGYCCAGGCTG
TTATGCAAGC-- |
|


|
|
YML081W(MacIsaac)/Yeast
| Match Rank: | 5 |
| Score: | 0.57
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | TTGYCCAGGCTG--
----CCAGTCTGAA |
|


|
|
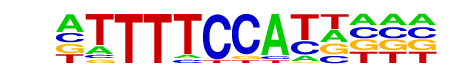
NFAT5/MA0606.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.57
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TTGYCCAGGCTG
ATTTTCCATT--- |
|

|
|
slbo/dmmpmm(Pollard)/fly
| Match Rank: | 7 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TTGYCCAGGCTG
ATTGCAAA----- |
|


|
|
slbo/MA0244.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TTGYCCAGGCTG
ATTGCAAA----- |
|


|
|
INO4/INO4_YPD/4-INO4,37-INO2(Harbison)/Yeast
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TTGYCCAGGCTG
TTTTCACATG--- |
|


|
|
WRKY62/MA1091.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.55
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TTGYCCAGGCTG
GTTGACCA----- |
|


|
|





















