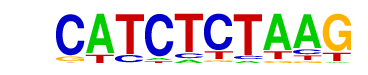
| p-value: | 1e-13 |
| log p-value: | -3.026e+01 |
| Information Content per bp: | 1.691 |
| Number of Target Sequences with motif | 87.0 |
| Percentage of Target Sequences with motif | 5.58% |
| Number of Background Sequences with motif | 15.1 |
| Percentage of Background Sequences with motif | 0.99% |
| Average Position of motif in Targets | 40.4 +/- 19.6bp |
| Average Position of motif in Background | 33.8 +/- 24.4bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.02 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Unknown2/Drosophila-Promoters/Homer
| Match Rank: | 1 |
| Score: | 0.77
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CTTAGAGATG
--TAGKGATG |
|


|
|
pros/dmmpmm(Bergman)/fly
| Match Rank: | 2 |
| Score: | 0.71
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CTTAGAGATG
---AGNCATG |
|


|
|
Tal1
| Match Rank: | 3 |
| Score: | 0.63
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | CTTAGAGATG
----CAGATG |
|


|
|
XBP1/Literature(Harbison)/Yeast
| Match Rank: | 4 |
| Score: | 0.62
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CTTAGAGATG
CTTCGAG--- |
|


|
|
TOD6?/SacCer-Promoters/Homer
| Match Rank: | 5 |
| Score: | 0.58
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | CTTAGAGATG----
----GCGATGAGMT |
|


|
|
PB0160.1_Rfxdc2_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.57
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CTTAGAGATG----
CTACTTGGATACGGAAT |
|


|
|
PRDM14(Zf)/H1-PRDM14-ChIP-Seq(GSE22767)/Homer
| Match Rank: | 7 |
| Score: | 0.57
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CTTAGAGATG-
GGTTAGAGACCT |
|


|
|
ZBTB18/MA0698.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.57
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CTTAGAGATG---
CATCCAGATGTTC |
|


|
|
NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer
| Match Rank: | 9 |
| Score: | 0.57
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CTTAGAGATG--
--AACAGATGGC |
|


|
|
THI2/THI2_Thi-/[](Harbison)/Yeast
| Match Rank: | 10 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CTTAGAGATG-
TCTTAGGGTTTC |
|


|
|