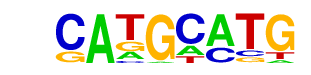
| p-value: | 1e-11 |
| log p-value: | -2.569e+01 |
| Information Content per bp: | 1.680 |
| Number of Target Sequences with motif | 185.0 |
| Percentage of Target Sequences with motif | 11.87% |
| Number of Background Sequences with motif | 78.8 |
| Percentage of Background Sequences with motif | 5.16% |
| Average Position of motif in Targets | 37.2 +/- 18.9bp |
| Average Position of motif in Background | 36.9 +/- 19.9bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.07 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
RBFox2(?)/Heart-RBFox2-CLIP-Seq(GSE57926)/Homer
| Match Rank: | 1 |
| Score: | 0.76
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CATGCATG
TGCATGCA-- |
|


|
|
LEC2/MA0581.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.73
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CATGCATG-
TGCATGCACAT |
|


|
|
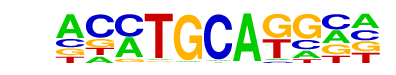
SOK2/MA0385.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.72
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CATGCATG--
ACCTGCAGGCA |
|

|
|
ABI3/MA0564.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.72
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CATGCATG
CTGCATGCA-- |
|


|
|
AtLEC2(ABI3/VP1)/Arabidopsis thaliana/AthaMap
| Match Rank: | 5 |
| Score: | 0.71
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CATGCATG
TCCATGCAAA |
|


|
|
PB0178.1_Sox8_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.70
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----CATGCATG-
NNTNTCATGAATGT |
|


|
|
PL0008.1_hlh-29/Jaspar
| Match Rank: | 7 |
| Score: | 0.69
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----CATGCATG-----
CTGCCACGCGTGGCCAA |
|


|
|
unc-86/MA0926.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.69
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CATGCATG-
-ATGCATAT |
|


|
|
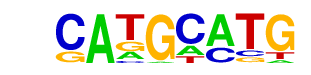
YAP5(MacIsaac)/Yeast
| Match Rank: | 9 |
| Score: | 0.67
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CATGCATG
-AAGCAT- |
|

|
|
Pax8(Paired,Homeobox)/Thyroid-Pax8-ChIP-Seq(GSE26938)/Homer
| Match Rank: | 10 |
| Score: | 0.67
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----CATGCATG--
SCAGYCADGCATGAC |
|


|
|