| p-value: | 1e-9 |
| log p-value: | -2.261e+01 |
| Information Content per bp: | 1.816 |
| Number of Target Sequences with motif | 45.0 |
| Percentage of Target Sequences with motif | 1.87% |
| Number of Background Sequences with motif | 5.0 |
| Percentage of Background Sequences with motif | 0.20% |
| Average Position of motif in Targets | 37.2 +/- 19.5bp |
| Average Position of motif in Background | 46.9 +/- 8.0bp |
| Strand Bias (log2 ratio + to - strand density) | 0.9 |
| Multiplicity (# of sites on avg that occur together) | 1.02 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0154.1_Osr1_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.74
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CATGYTACTT-----
ACATGCTACCTAATAC |
|


|
|
PB0155.1_Osr2_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.71
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CATGYTACTT-----
ACTTGCTACCTACACC |
|


|
|
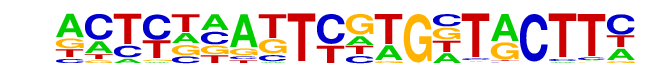
Mouse_Recombination_Hotspot(Zf)/Testis-DMC1-ChIP-Seq(GSE24438)/Homer
| Match Rank: | 3 |
| Score: | 0.69
| | Offset: | -9
| | Orientation: | forward strand |
| Alignment: | ---------CATGYTACTT-
ACTYKNATTCGTGNTACTTC |
|

|
|
PRDM9(Zf)/Testis-DMC1-ChIP-Seq(GSE35498)/Homer
| Match Rank: | 4 |
| Score: | 0.66
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CATGYTACTT----
AGATGCTRCTRCCHT |
|


|
|
odd/dmmpmm(Noyes)/fly
| Match Rank: | 5 |
| Score: | 0.66
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CATGYTACTT--
--TGCTACTGTT |
|


|
|
DMRT3/MA0610.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CATGYTACTT-
NTTGATACATT |
|


|
|
GAT3(MacIsaac)/Yeast
| Match Rank: | 7 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CATGYTACTT
CATGTTAT-- |
|


|
|
ovo/dmmpmm(Down)/fly
| Match Rank: | 8 |
| Score: | 0.63
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CATGYTACTT--
-CCGTTACTTTT |
|


|
|
LHY(Myb)/Seedling-LHY-ChIP-Seq(GSE52175)/Homer
| Match Rank: | 9 |
| Score: | 0.62
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CATGYTACTT--
--AGATATTTHT |
|


|
|
Brn1(POU,Homeobox)/NPC-Brn1-ChIP-Seq(GSE35496)/Homer
| Match Rank: | 10 |
| Score: | 0.62
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CATGYTACTT--
TATGCWAATBAV |
|


|
|





















