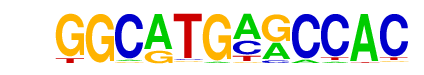
| p-value: | 1e-15 |
| log p-value: | -3.534e+01 |
| Information Content per bp: | 1.775 |
| Number of Target Sequences with motif | 88.0 |
| Percentage of Target Sequences with motif | 3.66% |
| Number of Background Sequences with motif | 13.9 |
| Percentage of Background Sequences with motif | 0.56% |
| Average Position of motif in Targets | 35.1 +/- 16.5bp |
| Average Position of motif in Background | 33.3 +/- 25.0bp |
| Strand Bias (log2 ratio + to - strand density) | -0.5 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
HES7/MA0822.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.68
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GTGGCTCATGCC-
-TGGCACGTGCCA |
|


|
|
HLHm5/dmmpmm(Pollard)/fly
| Match Rank: | 2 |
| Score: | 0.65
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GTGGCTCATGCC
-TGNCNCGTGC- |
|


|
|
E(spl)/dmmpmm(Pollard)/fly
| Match Rank: | 3 |
| Score: | 0.64
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GTGGCTCATGCC
TGTGCCACGTGC- |
|


|
|
HES5/MA0821.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.64
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GTGGCTCATGCC-
-TGGCACGTGCCG |
|


|
|
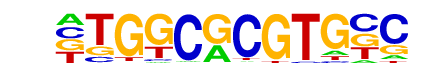
h/dmmpmm(Noyes)/fly
| Match Rank: | 5 |
| Score: | 0.64
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GTGGCTCATGCC
-TGGCGCGTGNC |
|

|
|
FUS3/MA0565.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.63
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | GTGGCTCATGCC
---GCGCATGCG |
|


|
|
BHLH104/MA0960.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.63
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | GTGGCTCATGCC
--GGCACGTGCC |
|


|
|
hkb/MA0450.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.62
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | GTGGCTCATGCC--
-----TCACGCCCC |
|


|
|
h/MA0449.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.62
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | GTGGCTCATGCC
--GGCACGTGCC |
|


|
|
RBFox2(?)/Heart-RBFox2-CLIP-Seq(GSE57926)/Homer
| Match Rank: | 10 |
| Score: | 0.62
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | GTGGCTCATGCC
----TGCATGCA |
|


|
|