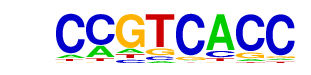
| p-value: | 1e-8 |
| log p-value: | -2.038e+01 |
| Information Content per bp: | 1.608 |
| Number of Target Sequences with motif | 78.0 |
| Percentage of Target Sequences with motif | 8.68% |
| Number of Background Sequences with motif | 19.5 |
| Percentage of Background Sequences with motif | 2.31% |
| Average Position of motif in Targets | 36.9 +/- 20.5bp |
| Average Position of motif in Background | 40.3 +/- 24.1bp |
| Strand Bias (log2 ratio + to - strand density) | 0.7 |
| Multiplicity (# of sites on avg that occur together) | 1.10 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
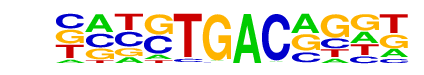
HVH21(HD-KNOTTED)/Hordeum vulgare/AthaMap
| Match Rank: | 1 |
| Score: | 0.72
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GGTGACGG--
CATGTGACAGGT |
|

|
|
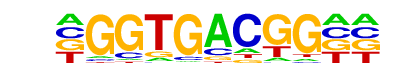
TGA1/MA0588.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.72
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GGTGACGG--
TGGTGACGTAA |
|

|
|
RPN4/MA0373.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.71
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GGTGACGG
GGTGGCG- |
|


|
|
TGA1(bZIP)/Arabidopsis thaliana/AthaMap
| Match Rank: | 4 |
| Score: | 0.69
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GGTGACGG--
TTGATGACGTTA |
|


|
|
hth/MA0227.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.68
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | GGTGACGG
--TGACAG |
|


|
|
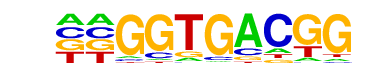
LEU3/MA0324.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.68
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GGTGACGG
CCGGTAACGG |
|

|
|
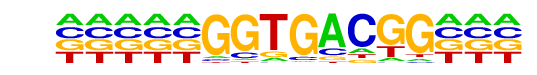
PB0117.1_Eomes_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.68
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----GGTGACGG---
GCGGAGGTGTCGCCTC |
|

|
|
Atf3/MA0605.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.67
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GGTGACGG
GATGACGT |
|


|
|
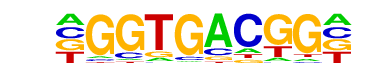
Hth/dmmpmm(Noyes_hd)/fly
| Match Rank: | 9 |
| Score: | 0.67
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GGTGACGG-
CTNTGACAGN |
|

|
|
TGA7/MA1070.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.67
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GGTGACGG--
GGTGACGTCA |
|


|
|