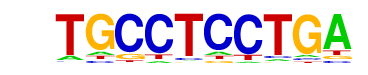
| p-value: | 1e-5 |
| log p-value: | -1.356e+01 |
| Information Content per bp: | 1.659 |
| Number of Target Sequences with motif | 92.0 |
| Percentage of Target Sequences with motif | 6.30% |
| Number of Background Sequences with motif | 42.8 |
| Percentage of Background Sequences with motif | 2.77% |
| Average Position of motif in Targets | 35.8 +/- 19.7bp |
| Average Position of motif in Background | 36.3 +/- 19.3bp |
| Strand Bias (log2 ratio + to - strand density) | -0.3 |
| Multiplicity (# of sites on avg that occur together) | 1.09 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
USV1/MA0413.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.66
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TGCCTCCTGA-------
AAATTCCCCCTGAATTTGTG |
|


|
|
E(spl)/dmmpmm(Papatsenko)/fly
| Match Rank: | 2 |
| Score: | 0.66
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TGCCTCCTGA
CGTGCCNNGTG- |
|


|
|
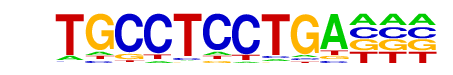
RAV1(2)(AP2/EREBP)/Arabidopsis thaliana/AthaMap
| Match Rank: | 3 |
| Score: | 0.65
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TGCCTCCTGA---
-ATCACCTGAGGC |
|

|
|
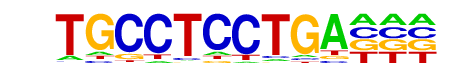
RAV1(var.2)/MA0583.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.65
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TGCCTCCTGA---
-ATCACCTGAGGC |
|

|
|
SOK2(MacIsaac)/Yeast
| Match Rank: | 5 |
| Score: | 0.65
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TGCCTCCTGA
TTGCCTGC--- |
|


|
|
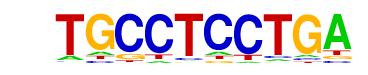
REI1/MA0364.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.65
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | TGCCTCCTGA
---CCCCTGA |
|

|
|
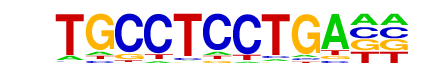
Zic(Zf)/Cerebellum-ZIC1.2-ChIP-Seq(GSE60731)/Homer
| Match Rank: | 7 |
| Score: | 0.64
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | TGCCTCCTGA--
--CCTGCTGAGH |
|

|
|
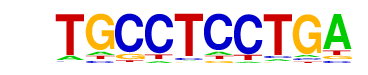
ZmHOX2a(1)(HD-HOX)/Zea mays/AthaMap
| Match Rank: | 8 |
| Score: | 0.64
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | TGCCTCCTGA
---GTCCTAA |
|

|
|
PHD1(MacIsaac)/Yeast
| Match Rank: | 9 |
| Score: | 0.64
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TGCCTCCTGA
GTGCCT----- |
|


|
|
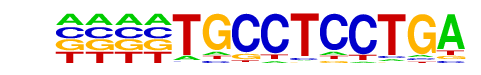
ETS:E-box(ETS,bHLH)/HPC7-Scl-ChIP-Seq(GSE22178)/Homer
| Match Rank: | 10 |
| Score: | 0.64
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----TGCCTCCTGA
CAGCTGTTTCCT-- |
|

|
|