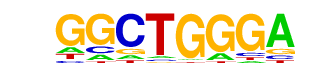







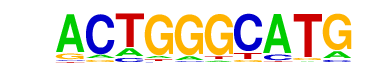



| p-value: | 1e-29 |
| log p-value: | -6.699e+01 |
| Information Content per bp: | 1.612 |
| Number of Target Sequences with motif | 360.0 |
| Percentage of Target Sequences with motif | 24.71% |
| Number of Background Sequences with motif | 134.1 |
| Percentage of Background Sequences with motif | 9.20% |
| Average Position of motif in Targets | 37.5 +/- 20.0bp |
| Average Position of motif in Background | 37.0 +/- 20.8bp |
| Strand Bias (log2 ratio + to - strand density) | -0.3 |
| Multiplicity (# of sites on avg that occur together) | 1.08 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.916 |  | 1e-25 | -59.113299 | 37.75% | 20.07% | motif file (matrix) |
| 2 | 0.832 |  | 1e-20 | -47.726119 | 15.24% | 4.88% | motif file (matrix) |
| 3 | 0.670 |  | 1e-19 | -44.318300 | 14.96% | 5.08% | motif file (matrix) |
| 4 | 0.818 |  | 1e-19 | -44.199686 | 9.47% | 1.96% | motif file (matrix) |
| 5 | 0.603 |  | 1e-11 | -27.060154 | 7.34% | 1.99% | motif file (matrix) |
| 6 | 0.673 |  | 1e-11 | -26.960205 | 3.64% | 0.30% | motif file (matrix) |
| 7 | 0.610 |  | 1e-9 | -21.458570 | 3.57% | 0.51% | motif file (matrix) |
| 8 | 0.606 |  | 1e-8 | -20.241974 | 1.99% | 0.03% | motif file (matrix) |
| 9 | 0.626 |  | 1e-7 | -16.970698 | 2.54% | 0.33% | motif file (matrix) |
| 10 | 0.623 |  | 1e-7 | -16.393673 | 5.28% | 1.78% | motif file (matrix) |