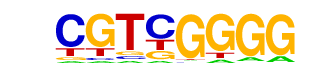
| p-value: | 1e-5 |
| log p-value: | -1.376e+01 |
| Information Content per bp: | 1.624 |
| Number of Target Sequences with motif | 34.0 |
| Percentage of Target Sequences with motif | 2.33% |
| Number of Background Sequences with motif | 5.7 |
| Percentage of Background Sequences with motif | 0.39% |
| Average Position of motif in Targets | 32.8 +/- 19.9bp |
| Average Position of motif in Background | 35.4 +/- 13.2bp |
| Strand Bias (log2 ratio + to - strand density) | -0.6 |
| Multiplicity (# of sites on avg that occur together) | 1.03 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
RDS2/MA0362.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.82
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CCCCGACG
CCCCGAN- |
|


|
|
ADR1/MA0268.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.78
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CCCCGACG
ACCCCAC-- |
|


|
|
NHP10/MA0344.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.75
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CCCCGACG
TCCCCGGC- |
|


|
|
MIG1/MA0337.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.75
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CCCCGACG
CCCCCGC-- |
|


|
|
MIG2/MA0338.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.74
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CCCCGACG
CCCCGCA- |
|


|
|
MIG3/MA0339.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.74
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CCCCGACG
CCCCGCA- |
|


|
|
MSN2/MA0341.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.74
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CCCCGACG
CCCCT--- |
|


|
|
MZF1/MA0056.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.73
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CCCCGACG
TCCCCA--- |
|


|
|
ZNF740/MA0753.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.72
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---CCCCGACG
CCCCCCCCAC- |
|


|
|
YPR022C/MA0436.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.72
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CCCCGACG
-CCCCACG |
|


|
|