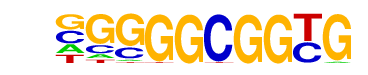| p-value: | 1e-5 |
| log p-value: | -1.365e+01 |
| Information Content per bp: | 1.658 |
| Number of Target Sequences with motif | 48.0 |
| Percentage of Target Sequences with motif | 2.73% |
| Number of Background Sequences with motif | 13.6 |
| Percentage of Background Sequences with motif | 0.73% |
| Average Position of motif in Targets | 40.7 +/- 17.9bp |
| Average Position of motif in Background | 36.1 +/- 20.8bp |
| Strand Bias (log2 ratio + to - strand density) | 0.8 |
| Multiplicity (# of sites on avg that occur together) | 1.12 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
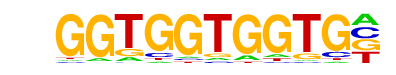
Run/dmmpmm(Papatsenko)/fly
| Match Rank: | 1 |
| Score: | 0.75
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GGTGGTGGTG
GGCGGTG--- |
|


|
|
ZBTB7C/MA0695.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.74
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GGTGGTGGTG
NTCGGTGGTCGC- |
|


|
|
ZBTB7B/MA0694.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.71
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GGTGGTGGTG
TTCGGTGGTCGC- |
|


|
|
ALFIN1(HD-PHD)/Medicago sativa/AthaMap
| Match Rank: | 4 |
| Score: | 0.68
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GGTGGTGGTG-
-GAGGTGGGGC |
|

|
|
ZBTB7A/MA0750.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.67
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GGTGGTGGTG
TCGGTGGTCGCN |
|


|
|
Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 6 |
| Score: | 0.67
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GGTGGTGGTG
GGGGGGGG-- |
|


|
|
ABI4(1)(AP2/EREBP)/Zea mays/AthaMap
| Match Rank: | 7 |
| Score: | 0.67
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GGTGGTGGTG
NGGGGCGGTG |
|

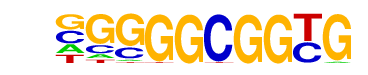
|
|
PB0196.1_Zbtb7b_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.66
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----GGTGGTGGTG--
NNANTGGTGGTCTTNNN |
|


|
|
EGR1/MA0162.2/Jaspar
| Match Rank: | 9 |
| Score: | 0.66
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GGTGGTGGTG-
GGCGGGGGCGGGGG |
|


|
|
ETS:RUNX(ETS,Runt)/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer
| Match Rank: | 10 |
| Score: | 0.66
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GGTGGTGGTG
ACAGGATGTGGT- |
|


|
|