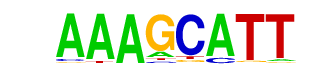
| p-value: | 1e-10 |
| log p-value: | -2.450e+01 |
| Information Content per bp: | 1.807 |
| Number of Target Sequences with motif | 159.0 |
| Percentage of Target Sequences with motif | 9.05% |
| Number of Background Sequences with motif | 69.6 |
| Percentage of Background Sequences with motif | 3.75% |
| Average Position of motif in Targets | 37.0 +/- 18.7bp |
| Average Position of motif in Background | 32.4 +/- 18.8bp |
| Strand Bias (log2 ratio + to - strand density) | 0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.02 |
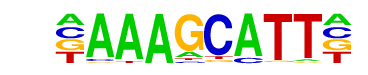
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
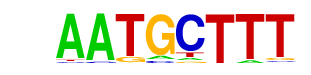
YAP5(MacIsaac)/Yeast
| Match Rank: | 1 |
| Score: | 0.82
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AAAGCATT
-AAGCAT- |
|

|
|
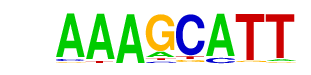
YAP5/MA0417.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.81
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AAAGCATT
-AAGCAT- |
|

|
|
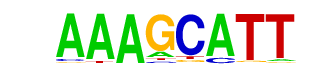
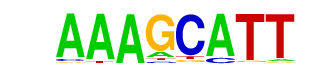
Dof2/MA0020.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.78
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AAAGCATT
AAAGCA-- |
|

|
|
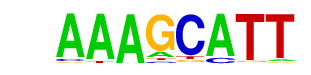
MNB1A/MA0053.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.75
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AAAGCATT
AAAGC--- |
|

|
|
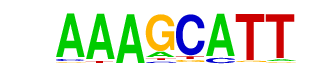
PBF/MA0064.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.75
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AAAGCATT
AAAGC--- |
|

|
|
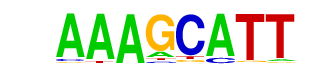
Dof3/MA0021.1/Jaspar
| Match Rank: | 6 |
| Score: | 0.74
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AAAGCATT
AAAGCG-- |
|

|
|
DOF2(C2C2(Zn) Dof)/Zea mays/AthaMap
| Match Rank: | 7 |
| Score: | 0.72
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----AAAGCATT
AAATAAAGCAA- |
|


|
|
HAT1/MA1024.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.69
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AAAGCATT-
CCAATCATTA |
|

|
|
PHYPADRAFT_140773/MA0987.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.68
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---AAAGCATT
CAAAAAGTAA- |
|


|
|
PHYPADRAFT_153324/MA0989.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.67
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---AAAGCATT
CAAAAAGTG-- |
|


|
|