




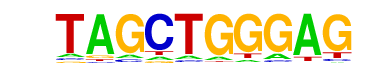




| p-value: | 1e-32 |
| log p-value: | -7.550e+01 |
| Information Content per bp: | 1.586 |
| Number of Target Sequences with motif | 319.0 |
| Percentage of Target Sequences with motif | 32.95% |
| Number of Background Sequences with motif | 94.8 |
| Percentage of Background Sequences with motif | 10.45% |
| Average Position of motif in Targets | 36.8 +/- 19.2bp |
| Average Position of motif in Background | 40.6 +/- 21.7bp |
| Strand Bias (log2 ratio + to - strand density) | 0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.20 |
| Motif File: | file (matrix) reverse opposite |
| Rank | Match Score | Redundant Motif | P-value | log P-value | % of Targets | % of Background | Motif file |
| 1 | 0.885 |  | 1e-30 | -69.579116 | 37.19% | 14.20% | motif file (matrix) |
| 2 | 0.698 |  | 1e-24 | -55.891320 | 26.45% | 8.70% | motif file (matrix) |
| 3 | 0.786 |  | 1e-24 | -55.755344 | 34.92% | 14.61% | motif file (matrix) |
| 4 | 0.731 |  | 1e-22 | -52.018028 | 13.84% | 1.99% | motif file (matrix) |
| 5 | 0.602 |  | 1e-19 | -45.778149 | 23.14% | 7.88% | motif file (matrix) |
| 6 | 0.698 |  | 1e-18 | -42.419237 | 10.64% | 1.41% | motif file (matrix) |
| 7 | 0.672 |  | 1e-15 | -35.973644 | 10.02% | 1.61% | motif file (matrix) |
| 8 | 0.671 |  | 1e-14 | -33.613704 | 14.26% | 4.02% | motif file (matrix) |