| p-value: | 1e-22 |
| log p-value: | -5.108e+01 |
| Information Content per bp: | 1.607 |
| Number of Target Sequences with motif | 213.0 |
| Percentage of Target Sequences with motif | 12.16% |
| Number of Background Sequences with motif | 64.1 |
| Percentage of Background Sequences with motif | 3.53% |
| Average Position of motif in Targets | 37.7 +/- 18.1bp |
| Average Position of motif in Background | 32.0 +/- 18.0bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.02 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
POL009.1_DCE_S_II/Jaspar
| Match Rank: | 1 |
| Score: | 0.69
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | SCTGGRATBMAA
GCTGTG------ |
|


|
|
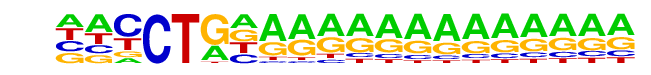
SA0002.1_at_AC_acceptor/Jaspar
| Match Rank: | 2 |
| Score: | 0.69
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --SCTGGRATBMAA------
NNCCTGNAAAAAAAAAAAAA |
|


|
|
SA0001.1_at_AC_acceptor/Jaspar
| Match Rank: | 3 |
| Score: | 0.68
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --SCTGGRATBMAA------
NNCCTGNAAAAAAAAAAAAA |
|

|
|
SWI5/Literature(Harbison)/Yeast
| Match Rank: | 4 |
| Score: | 0.66
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -SCTGGRATBMAA
GGCTGA------- |
|


|
|
POL002.1_INR/Jaspar
| Match Rank: | 5 |
| Score: | 0.63
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---SCTGGRATBMAA
NNNANTGA------- |
|


|
|
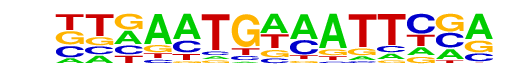
PB0169.1_Sox15_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.63
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---SCTGGRATBMAA
TTGAATGAAATTCGA |
|

|
|
TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 7 |
| Score: | 0.62
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | SCTGGRATBMAA
NCTGGAATGC-- |
|


|
|
RAV1(2)(AP2/EREBP)/Arabidopsis thaliana/AthaMap
| Match Rank: | 8 |
| Score: | 0.62
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----SCTGGRATBMAA
ATCACCTGAGGC---- |
|


|
|
RAV1(var.2)/MA0583.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.62
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----SCTGGRATBMAA
ATCACCTGAGGC---- |
|


|
|
TEAD2(TEA)/Py2T-Tead2-ChIP-Seq(GSE55709)/Homer
| Match Rank: | 10 |
| Score: | 0.62
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | SCTGGRATBMAA
CCWGGAATGY-- |
|


|
|





















