| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | PABPC1(?)/MEL-PABC1-CLIP-Seq(GSE69755)/Homer | 1e-7 | -1.689e+01 | 0.0000 | 297.0 | 16.96% | 195.7 | 10.79% | motif file (matrix) |
pdf |
| 2 |  | GSC(Homeobox)/FrogEmbryos-GSC-ChIP-Seq(DRA000576)/Homer | 1e-5 | -1.257e+01 | 0.0007 | 203.0 | 11.59% | 130.6 | 7.20% | motif file (matrix) |
pdf |
| 3 |  | SUT1?/SacCer-Promoters/Homer | 1e-4 | -1.151e+01 | 0.0013 | 414.0 | 23.64% | 323.0 | 17.80% | motif file (matrix) |
pdf |
| 4 |  | LHY(Myb)/Seedling-LHY-ChIP-Seq(GSE52175)/Homer | 1e-4 | -1.088e+01 | 0.0018 | 159.0 | 9.08% | 99.3 | 5.47% | motif file (matrix) |
pdf |
| 5 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-3 | -8.389e+00 | 0.0176 | 120.0 | 6.85% | 75.8 | 4.18% | motif file (matrix) |
pdf |
| 6 |  | DMRT1(DM)/Testis-DMRT1-ChIP-Seq(GSE64892)/Homer | 1e-3 | -8.301e+00 | 0.0176 | 34.0 | 1.94% | 11.4 | 0.63% | motif file (matrix) |
pdf |
| 7 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-3 | -7.907e+00 | 0.0203 | 136.0 | 7.77% | 90.7 | 5.00% | motif file (matrix) |
pdf |
| 8 | 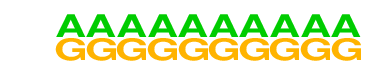 | SeqBias: G/A bias | 1e-3 | -7.845e+00 | 0.0203 | 1720.0 | 98.23% | 1748.9 | 96.41% | motif file (matrix) |
pdf |
| 9 |  | Reverb(NR),DR2/RAW-Reverba.biotin-ChIP-Seq(GSE45914)/Homer | 1e-3 | -7.837e+00 | 0.0203 | 11.0 | 0.63% | 0.9 | 0.05% | motif file (matrix) |
pdf |
| 10 |  | ZNF416(Zf)/HEK293-ZNF416.GFP-ChIP-Seq(GSE58341)/Homer | 1e-3 | -7.386e+00 | 0.0240 | 129.0 | 7.37% | 86.2 | 4.75% | motif file (matrix) |
pdf |
| 11 |  | CHR(?)/Hela-CellCycle-Expression/Homer | 1e-3 | -7.374e+00 | 0.0240 | 84.0 | 4.80% | 49.6 | 2.74% | motif file (matrix) |
pdf |
| 12 |  | SeqBias: TA-repeat | 1e-3 | -7.356e+00 | 0.0240 | 500.0 | 28.56% | 431.1 | 23.76% | motif file (matrix) |
pdf |
| 13 |  | Erra(NR)/HepG2-Erra-ChIP-Seq(GSE31477)/Homer | 1e-3 | -6.978e+00 | 0.0277 | 140.0 | 8.00% | 97.0 | 5.35% | motif file (matrix) |
pdf |
| 14 |  | DMRT6(DM)/Testis-DMRT6-ChIP-Seq(GSE60440)/Homer | 1e-3 | -6.908e+00 | 0.0277 | 26.0 | 1.48% | 8.4 | 0.46% | motif file (matrix) |
pdf |
| 15 |  | Mef2c(MADS)/GM12878-Mef2c-ChIP-Seq(GSE32465)/Homer | 1e-2 | -6.217e+00 | 0.0515 | 71.0 | 4.05% | 42.2 | 2.33% | motif file (matrix) |
pdf |
| 16 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-2 | -6.160e+00 | 0.0515 | 77.0 | 4.40% | 48.0 | 2.64% | motif file (matrix) |
pdf |
| 17 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-2 | -6.001e+00 | 0.0563 | 19.0 | 1.09% | 5.4 | 0.30% | motif file (matrix) |
pdf |
| 18 |  | ZNF519(Zf)/HEK293-ZNF519.GFP-ChIP-Seq(GSE58341)/Homer | 1e-2 | -5.823e+00 | 0.0636 | 15.0 | 0.86% | 3.0 | 0.17% | motif file (matrix) |
pdf |
| 19 |  | Usf2(bHLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-2 | -5.738e+00 | 0.0656 | 31.0 | 1.77% | 13.9 | 0.76% | motif file (matrix) |
pdf |
| 20 |  | Bapx1(Homeobox)/VertebralCol-Bapx1-ChIP-Seq(GSE36672)/Homer | 1e-2 | -5.111e+00 | 0.1167 | 203.0 | 11.59% | 163.5 | 9.01% | motif file (matrix) |
pdf |
| 21 |  | ZNF467(Zf)/HEK293-ZNF467.GFP-ChIP-Seq(GSE58341)/Homer | 1e-2 | -5.061e+00 | 0.1168 | 58.0 | 3.31% | 35.3 | 1.95% | motif file (matrix) |
pdf |
| 22 |  | PHA-4(Forkhead)/cElegans-Embryos-PHA4-ChIP-Seq(modEncode)/Homer | 1e-2 | -5.049e+00 | 0.1168 | 299.0 | 17.08% | 255.0 | 14.06% | motif file (matrix) |
pdf |
| 23 |  | Etv2(ETS)/ES-ER71-ChIP-Seq(GSE59402)/Homer(0.967) | 1e-2 | -4.925e+00 | 0.1222 | 50.0 | 2.86% | 29.5 | 1.63% | motif file (matrix) |
pdf |
| 24 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-2 | -4.702e+00 | 0.1464 | 31.0 | 1.77% | 15.7 | 0.86% | motif file (matrix) |
pdf |















































