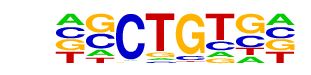
| p-value: | 1e-14 |
| log p-value: | -3.247e+01 |
| Information Content per bp: | 1.951 |
| Number of Target Sequences with motif | 139.0 |
| Percentage of Target Sequences with motif | 5.68% |
| Number of Background Sequences with motif | 42.6 |
| Percentage of Background Sequences with motif | 1.68% |
| Average Position of motif in Targets | 38.7 +/- 18.2bp |
| Average Position of motif in Background | 40.4 +/- 24.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.3 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
HMRA2/MA0318.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.74
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GCCTGTAA-
-CATGTAAT |
|


|
|
MATALPHA2/MA0328.2/Jaspar
| Match Rank: | 2 |
| Score: | 0.73
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GCCTGTAA-
-CGTGTAAT |
|


|
|
POL009.1_DCE_S_II/Jaspar
| Match Rank: | 3 |
| Score: | 0.67
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GCCTGTAA
-GCTGTG- |
|

|
|
SOK2(MacIsaac)/Yeast
| Match Rank: | 4 |
| Score: | 0.65
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GCCTGTAA
TTGCCTGC-- |
|


|
|
MEIS2/MA0774.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.65
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GCCTGTAA-
-GCTGTCAA |
|


|
|
PH0141.1_Pknox2/Jaspar
| Match Rank: | 6 |
| Score: | 0.65
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GCCTGTAA----
AAGCACCTGTCAATAT |
|


|
|
INO4/MA0322.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GCCTGTAA-
GCATGTGAA |
|


|
|
INO4(MacIsaac)/Yeast
| Match Rank: | 8 |
| Score: | 0.62
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GCCTGTAA-
GCATGTGAA |
|


|
|
achi/MA0207.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.62
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | GCCTGTAA
--CTGTCA |
|


|
|
ara/MA0210.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.62
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | GCCTGTAA
---TGTTA |
|


|
|





















