| p-value: | 1e-29 |
| log p-value: | -6.836e+01 |
| Information Content per bp: | 1.508 |
| Number of Target Sequences with motif | 231.0 |
| Percentage of Target Sequences with motif | 9.44% |
| Number of Background Sequences with motif | 54.4 |
| Percentage of Background Sequences with motif | 2.15% |
| Average Position of motif in Targets | 38.0 +/- 17.9bp |
| Average Position of motif in Background | 39.0 +/- 24.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.13 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
CRZ1(MacIsaac)/Yeast
| Match Rank: | 1 |
| Score: | 0.73
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CCTCAGCCTCCC
---CAGCCAC-- |
|


|
|
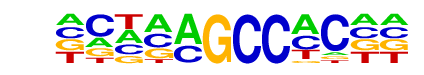
CRZ1/MA0285.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.65
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CCTCAGCCTCCC
-CTAAGCCAC-- |
|

|
|
bZIP910(2)(bZIP)/Antirrhinum majus/AthaMap
| Match Rank: | 3 |
| Score: | 0.62
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CCTCAGCCTCCC
ACGTCAGCACCC- |
|


|
|
ZNF519(Zf)/HEK293-ZNF519.GFP-ChIP-Seq(GSE58341)/Homer
| Match Rank: | 4 |
| Score: | 0.61
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CCTCAGCCTCCC
GCTCGGSCTC-- |
|


|
|
SWI5/Literature(Harbison)/Yeast
| Match Rank: | 5 |
| Score: | 0.61
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CCTCAGCCTCCC
--CCAGCA---- |
|


|
|
ETS:RUNX(ETS,Runt)/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer
| Match Rank: | 6 |
| Score: | 0.60
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CCTCAGCCTCCC
ACCACATCCTGT- |
|


|
|
ADR1/Literature(Harbison)/Yeast
| Match Rank: | 7 |
| Score: | 0.60
| | Offset: | 6
| | Orientation: | reverse strand |
| Alignment: | CCTCAGCCTCCC
------ACCCCN |
|


|
|
ADR1(MacIsaac)/Yeast
| Match Rank: | 8 |
| Score: | 0.60
| | Offset: | 6
| | Orientation: | reverse strand |
| Alignment: | CCTCAGCCTCCC
------ACCCCN |
|


|
|
PB0097.1_Zfp281_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.59
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CCTCAGCCTCCC--
TCCCCCCCCCCCCCC |
|


|
|
SP1/MA0079.3/Jaspar
| Match Rank: | 10 |
| Score: | 0.58
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CCTCAGCCTCCC
GCCCCGCCCCC- |
|


|
|





















