| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | PABPC1(?)/MEL-PABC1-CLIP-Seq(GSE69755)/Homer | 1e-11 | -2.744e+01 | 0.0000 | 435.0 | 17.77% | 274.1 | 10.81% | motif file (matrix) |
pdf |
| 2 |  | FoxL2(Forkhead)/Ovary-FoxL2-ChIP-Seq(GSE60858)/Homer | 1e-8 | -1.943e+01 | 0.0000 | 171.0 | 6.99% | 85.8 | 3.39% | motif file (matrix) |
pdf |
| 3 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-8 | -1.891e+01 | 0.0000 | 196.0 | 8.01% | 105.2 | 4.15% | motif file (matrix) |
pdf |
| 4 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-7 | -1.787e+01 | 0.0000 | 222.0 | 9.07% | 128.0 | 5.05% | motif file (matrix) |
pdf |
| 5 |  | SeqBias: TA-repeat | 1e-7 | -1.758e+01 | 0.0000 | 728.0 | 29.74% | 580.6 | 22.91% | motif file (matrix) |
pdf |
| 6 |  | PHA-4(Forkhead)/cElegans-Embryos-PHA4-ChIP-Seq(modEncode)/Homer | 1e-7 | -1.692e+01 | 0.0000 | 488.0 | 19.93% | 360.5 | 14.22% | motif file (matrix) |
pdf |
| 7 |  | FOXM1(Forkhead)/MCF7-FOXM1-ChIP-Seq(GSE72977)/Homer | 1e-5 | -1.313e+01 | 0.0001 | 192.0 | 7.84% | 119.0 | 4.70% | motif file (matrix) |
pdf |
| 8 |  | ZNF416(Zf)/HEK293-ZNF416.GFP-ChIP-Seq(GSE58341)/Homer | 1e-4 | -1.117e+01 | 0.0007 | 172.0 | 7.03% | 108.3 | 4.27% | motif file (matrix) |
pdf |
| 9 |  | Gfi1b(Zf)/HPC7-Gfi1b-ChIP-Seq(GSE22178)/Homer | 1e-4 | -9.605e+00 | 0.0029 | 105.0 | 4.29% | 59.6 | 2.35% | motif file (matrix) |
pdf |
| 10 |  | Oct6(POU,Homeobox)/NPC-Oct6-ChIP-Seq(GSE35496)/Homer | 1e-4 | -9.249e+00 | 0.0037 | 72.0 | 2.94% | 35.6 | 1.41% | motif file (matrix) |
pdf |
| 11 |  | Brn1(POU,Homeobox)/NPC-Brn1-ChIP-Seq(GSE35496)/Homer | 1e-3 | -8.411e+00 | 0.0078 | 57.0 | 2.33% | 26.0 | 1.03% | motif file (matrix) |
pdf |
| 12 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-3 | -8.207e+00 | 0.0088 | 151.0 | 6.17% | 101.8 | 4.02% | motif file (matrix) |
pdf |
| 13 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-3 | -7.661e+00 | 0.0140 | 59.0 | 2.41% | 29.0 | 1.15% | motif file (matrix) |
pdf |
| 14 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.644e+00 | 0.0140 | 139.0 | 5.68% | 93.3 | 3.68% | motif file (matrix) |
pdf |
| 15 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-3 | -7.222e+00 | 0.0189 | 67.0 | 2.74% | 36.8 | 1.45% | motif file (matrix) |
pdf |
| 16 |  | IBL1(bHLH)/Seedling-IBL1-ChIP-Seq(GSE51120)/Homer | 1e-3 | -7.132e+00 | 0.0193 | 146.0 | 5.96% | 101.9 | 4.02% | motif file (matrix) |
pdf |
| 17 |  | Srebp2(bHLH)/HepG2-Srebp2-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.099e+00 | 0.0193 | 23.0 | 0.94% | 7.0 | 0.28% | motif file (matrix) |
pdf |
| 18 |  | FOXA1:AR(Forkhead,NR)/LNCAP-AR-ChIP-Seq(GSE27824)/Homer | 1e-3 | -6.994e+00 | 0.0197 | 15.0 | 0.61% | 2.4 | 0.09% | motif file (matrix) |
pdf |
| 19 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan_et_al.)/Homer | 1e-2 | -6.902e+00 | 0.0205 | 204.0 | 8.33% | 153.6 | 6.06% | motif file (matrix) |
pdf |
| 20 | 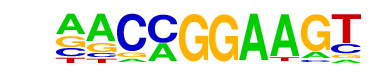 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-2 | -6.752e+00 | 0.0226 | 98.0 | 4.00% | 62.8 | 2.48% | motif file (matrix) |
pdf |
| 21 |  | Usf2(bHLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-2 | -6.398e+00 | 0.0307 | 47.0 | 1.92% | 23.9 | 0.94% | motif file (matrix) |
pdf |
| 22 |  | Unknown6/Drosophila-Promoters/Homer | 1e-2 | -6.317e+00 | 0.0318 | 150.0 | 6.13% | 108.7 | 4.29% | motif file (matrix) |
pdf |
| 23 |  | PIF4(bHLH)/Seedling-PIF4-ChIP-Seq(GSE35315)/Homer | 1e-2 | -6.228e+00 | 0.0332 | 130.0 | 5.31% | 91.4 | 3.61% | motif file (matrix) |
pdf |
| 24 |  | bHLHE40(bHLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.105e+00 | 0.0360 | 32.0 | 1.31% | 13.7 | 0.54% | motif file (matrix) |
pdf |
| 25 |  | IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer | 1e-2 | -6.065e+00 | 0.0360 | 54.0 | 2.21% | 29.8 | 1.18% | motif file (matrix) |
pdf |
| 26 |  | PU.1:IRF8(ETS:IRF)/pDC-Irf8-ChIP-Seq(GSE66899)/Homer | 1e-2 | -5.914e+00 | 0.0402 | 27.0 | 1.10% | 10.6 | 0.42% | motif file (matrix) |
pdf |
| 27 |  | E-box/Arabidopsis-Promoters/Homer | 1e-2 | -5.890e+00 | 0.0402 | 40.0 | 1.63% | 19.3 | 0.76% | motif file (matrix) |
pdf |
| 28 |  | HIF-1b(HLH)/T47D-HIF1b-ChIP-Seq(GSE59937)/Homer | 1e-2 | -5.870e+00 | 0.0402 | 81.0 | 3.31% | 51.1 | 2.02% | motif file (matrix) |
pdf |
| 29 |  | c-Myc(bHLH)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.855e+00 | 0.0402 | 52.0 | 2.12% | 28.4 | 1.12% | motif file (matrix) |
pdf |
| 30 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-2 | -5.641e+00 | 0.0458 | 98.0 | 4.00% | 66.9 | 2.64% | motif file (matrix) |
pdf |
| 31 |  | Oct4(POU,Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.593e+00 | 0.0465 | 75.0 | 3.06% | 47.3 | 1.87% | motif file (matrix) |
pdf |
| 32 |  | GATA:SCL(Zf,bHLH)/Ter119-SCL-ChIP-Seq(GSE18720)/Homer | 1e-2 | -5.230e+00 | 0.0647 | 12.0 | 0.49% | 2.9 | 0.11% | motif file (matrix) |
pdf |
| 33 |  | Pho2(bHLH)/Yeast-Pho2-ChIP-Seq(GSE29506)/Homer | 1e-2 | -4.816e+00 | 0.0950 | 30.0 | 1.23% | 14.8 | 0.58% | motif file (matrix) |
pdf |
| 34 |  | Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer | 1e-2 | -4.767e+00 | 0.0969 | 119.0 | 4.86% | 88.2 | 3.48% | motif file (matrix) |
pdf |
| 35 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-2 | -4.761e+00 | 0.0969 | 57.0 | 2.33% | 35.6 | 1.40% | motif file (matrix) |
pdf |
| 36 |  | AR-halfsite(NR)/LNCaP-AR-ChIP-Seq(GSE27824)/Homer | 1e-2 | -4.728e+00 | 0.0969 | 344.0 | 14.05% | 298.2 | 11.77% | motif file (matrix) |
pdf |
| 37 |  | SeqBias: GA-repeat | 1e-2 | -4.704e+00 | 0.0969 | 851.0 | 34.76% | 800.0 | 31.57% | motif file (matrix) |
pdf |
| 38 |  | GAGA-repeat/SacCer-Promoters/Homer | 1e-2 | -4.669e+00 | 0.0969 | 514.0 | 21.00% | 464.3 | 18.32% | motif file (matrix) |
pdf |
| 39 |  | Bapx1(Homeobox)/VertebralCol-Bapx1-ChIP-Seq(GSE36672)/Homer | 1e-2 | -4.634e+00 | 0.0969 | 254.0 | 10.38% | 213.4 | 8.42% | motif file (matrix) |
pdf |













































































