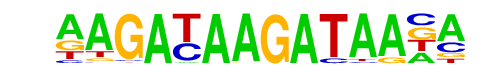| p-value: | 1e-7 |
| log p-value: | -1.721e+01 |
| Information Content per bp: | 1.784 |
| Number of Target Sequences with motif | 24.0 |
| Percentage of Target Sequences with motif | 0.63% |
| Number of Background Sequences with motif | 0.9 |
| Percentage of Background Sequences with motif | 0.02% |
| Average Position of motif in Targets | 43.3 +/- 17.6bp |
| Average Position of motif in Background | 66.0 +/- 0.0bp |
| Strand Bias (log2 ratio + to - strand density) | 1.3 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Mecom/MA0029.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.71
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GAAAAGAAAACA
AAGATAAGATAACA |
|

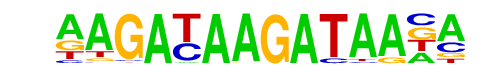
|
|
AZF1/MA0277.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.71
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GAAAAGAAAACA
AAAAAGAAA--- |
|


|
|
GLN3/GLN3_RAPA/8-GZF3,11-GLN3(Harbison)/Yeast
| Match Rank: | 3 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GAAAAGAAAACA
GATAAGATA--- |
|


|
|
FKH2/FKH2_YPD/18-FKH1,18-FKH2(Harbison)/Yeast
| Match Rank: | 4 |
| Score: | 0.63
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GAAAAGAAAACA-
-AAANGTAAACAA |
|


|
|
DAL82/DAL82_SM/3-DAL82(Harbison)/Yeast
| Match Rank: | 5 |
| Score: | 0.63
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GAAAAGAAAACA
GATAAGA----- |
|


|
|
PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 6 |
| Score: | 0.62
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GAAAAGAAAACA
CGGAAGTGAAAC-- |
|


|
|
GLN3(MacIsaac)/Yeast
| Match Rank: | 7 |
| Score: | 0.60
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GAAAAGAAAACA
AGATAAGATA--- |
|


|
|
FOXP1/MA0481.1/Jaspar
| Match Rank: | 8 |
| Score: | 0.60
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GAAAAGAAAACA---
CAAAAGTAAACAAAG |
|


|
|
GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer
| Match Rank: | 9 |
| Score: | 0.60
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | GAAAAGAAAACA
----AGATAASR |
|


|
|
ZNF189(Zf)/HEK293-ZNF189.GFP-ChIP-Seq(GSE58341)/Homer
| Match Rank: | 10 |
| Score: | 0.59
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GAAAAGAAAACA
TGGAACAGMA---- |
|


|
|