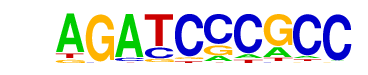
| p-value: | 1e-5 |
| log p-value: | -1.308e+01 |
| Information Content per bp: | 1.700 |
| Number of Target Sequences with motif | 27.0 |
| Percentage of Target Sequences with motif | 1.77% |
| Number of Background Sequences with motif | 2.9 |
| Percentage of Background Sequences with motif | 0.21% |
| Average Position of motif in Targets | 48.3 +/- 15.8bp |
| Average Position of motif in Background | 44.3 +/- 9.8bp |
| Strand Bias (log2 ratio + to - strand density) | 0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
POL011.1_XCPE1/Jaspar
| Match Rank: | 1 |
| Score: | 0.71
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | AGATCCCGCC-
-GGTCCCGCCC |
|


|
|
UGA3/MA0410.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.71
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | AGATCCCGCC-
---TCCCGCCG |
|


|
|
ECM23/MA0293.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.67
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AGATCCCGCC
AAAGATCTAAA- |
|


|
|
PB0039.1_Klf7_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.67
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AGATCCCGCC-----
TCGACCCCGCCCCTAT |
|


|
|
GAT4/MA0302.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.66
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AGATCCCGCC
AAAGATCTAAA- |
|


|
|
ZmHOX2a(2)(HD-HOX)/Zea mays/AthaMap
| Match Rank: | 6 |
| Score: | 0.66
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AGATCCCGCC
CAGATCA---- |
|


|
|
E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer
| Match Rank: | 7 |
| Score: | 0.66
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | AGATCCCGCC--
--TTCCCGCCWG |
|


|
|
POL006.1_BREu/Jaspar
| Match Rank: | 8 |
| Score: | 0.66
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | AGATCCCGCC
--AGCGCGCC |
|

|
|
SRD1/MA0389.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.65
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AGATCCCGCC
GTAGATCT---- |
|


|
|
PB0164.1_Smad3_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.65
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AGATCCCGCC------
TACGCCCCGCCACTCTG |
|


|
|