| p-value: | 1e-8 |
| log p-value: | -2.023e+01 |
| Information Content per bp: | 1.678 |
| Number of Target Sequences with motif | 59.0 |
| Percentage of Target Sequences with motif | 1.55% |
| Number of Background Sequences with motif | 12.9 |
| Percentage of Background Sequences with motif | 0.32% |
| Average Position of motif in Targets | 35.1 +/- 18.0bp |
| Average Position of motif in Background | 25.3 +/- 13.1bp |
| Strand Bias (log2 ratio + to - strand density) | 0.7 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
achi/MA0207.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.84
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | CGCTCTGTCA
----CTGTCA |
|


|
|
hth/MA0227.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.82
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | CGCTCTGTCA
----CTGTCA |
|


|
|
vis/MA0252.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.82
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | CGCTCTGTCA
----CTGTCA |
|


|
|
MEIS2/MA0774.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.80
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CGCTCTGTCA-
---GCTGTCAA |
|


|
|
MEIS1/MA0498.2/Jaspar
| Match Rank: | 5 |
| Score: | 0.80
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | CGCTCTGTCA-
----NTGTCAN |
|


|
|
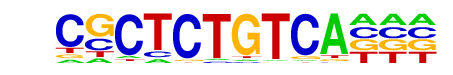
Hth/dmmpmm(Noyes_hd)/fly
| Match Rank: | 6 |
| Score: | 0.80
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CGCTCTGTCA---
---NCTGTCANAG |
|

|
|
MEIS3/MA0775.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.79
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CGCTCTGTCA-
---CCTGTCAA |
|


|
|
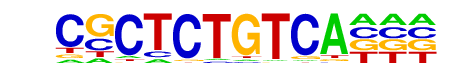
Vis/dmmpmm(Noyes_hd)/fly
| Match Rank: | 8 |
| Score: | 0.78
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CGCTCTGTCA---
---NCTGTCANNT |
|

|
|
TOS8/MA0408.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.77
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | CGCTCTGTCA--
----CTGTCAAA |
|


|
|
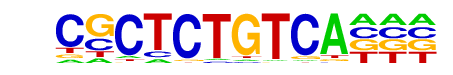
Achi/dmmpmm(Noyes_hd)/fly
| Match Rank: | 10 |
| Score: | 0.77
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CGCTCTGTCA---
---GCTGTCAAAN |
|

|
|





















