| p-value: | 1e-31 |
| log p-value: | -7.301e+01 |
| Information Content per bp: | 1.606 |
| Number of Target Sequences with motif | 1391.0 |
| Percentage of Target Sequences with motif | 36.44% |
| Number of Background Sequences with motif | 973.8 |
| Percentage of Background Sequences with motif | 24.21% |
| Average Position of motif in Targets | 37.2 +/- 20.2bp |
| Average Position of motif in Background | 36.4 +/- 19.7bp |
| Strand Bias (log2 ratio + to - strand density) | -0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.29 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
RME1(MacIsaac)/Yeast
| Match Rank: | 1 |
| Score: | 0.78
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TTTCTTTG--
TTCCTTTGGA |
|


|
|
RME1/MA0370.1/Jaspar
| Match Rank: | 2 |
| Score: | 0.77
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TTTCTTTG--
TTCCTTTGGA |
|


|
|
AZF1/MA0277.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.76
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TTTCTTTG-
TTTCTTTTT |
|


|
|
Dof2/MA0020.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.75
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TTTCTTTG
-NGCTTT- |
|


|
|
MNB1A/MA0053.1/Jaspar
| Match Rank: | 5 |
| Score: | 0.75
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TTTCTTTG
--GCTTT- |
|


|
|
PB0083.1_Tcf7_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.73
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TTTCTTTG-------
NNTTCCTTTGATCTNNA |
|


|
|
PBF/MA0064.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.73
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TTTCTTTG
--GCTTT- |
|


|
|
PB0166.1_Sox12_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.73
| | Offset: | -1
| | Orientation: | reverse strand |
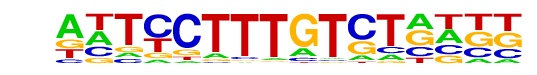
| Alignment: | -TTTCTTTG-------
ANTCCTTTGTCTNNNN |
|

|
|
DOF2(C2C2(Zn) Dof)/Zea mays/AthaMap
| Match Rank: | 9 |
| Score: | 0.73
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TTTCTTTG---
NNGCTTTANNN |
|


|
|
pan/dmmpmm(Papatsenko)/fly
| Match Rank: | 10 |
| Score: | 0.72
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | TTTCTTTG--
---CTTTGAT |
|


|
|





















