| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | ZNF416(Zf)/HEK293-ZNF416.GFP-ChIP-Seq(GSE58341)/Homer | 1e-14 | -3.234e+01 | 0.0000 | 298.0 | 7.81% | 152.6 | 3.80% | motif file (matrix) |
pdf |
| 2 |  | PABPC1(?)/MEL-PABC1-CLIP-Seq(GSE69755)/Homer | 1e-11 | -2.588e+01 | 0.0000 | 643.0 | 16.85% | 462.8 | 11.51% | motif file (matrix) |
pdf |
| 3 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-10 | -2.526e+01 | 0.0000 | 341.0 | 8.93% | 204.2 | 5.08% | motif file (matrix) |
pdf |
| 4 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-10 | -2.412e+01 | 0.0000 | 296.0 | 7.75% | 171.6 | 4.27% | motif file (matrix) |
pdf |
| 5 |  | FoxL2(Forkhead)/Ovary-FoxL2-ChIP-Seq(GSE60858)/Homer | 1e-10 | -2.398e+01 | 0.0000 | 260.0 | 6.81% | 143.3 | 3.56% | motif file (matrix) |
pdf |
| 6 |  | PHA-4(Forkhead)/cElegans-Embryos-PHA4-ChIP-Seq(modEncode)/Homer | 1e-9 | -2.110e+01 | 0.0000 | 756.0 | 19.81% | 588.1 | 14.62% | motif file (matrix) |
pdf |
| 7 |  | FOXM1(Forkhead)/MCF7-FOXM1-ChIP-Seq(GSE72977)/Homer | 1e-9 | -2.080e+01 | 0.0000 | 273.0 | 7.15% | 162.7 | 4.04% | motif file (matrix) |
pdf |
| 8 |  | SeqBias: TA-repeat | 1e-8 | -1.933e+01 | 0.0000 | 1115.0 | 29.21% | 943.8 | 23.47% | motif file (matrix) |
pdf |
| 9 |  | GSC(Homeobox)/FrogEmbryos-GSC-ChIP-Seq(DRA000576)/Homer | 1e-6 | -1.413e+01 | 0.0000 | 410.0 | 10.74% | 305.0 | 7.58% | motif file (matrix) |
pdf |
| 10 |  | NPAS2(bHLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer | 1e-5 | -1.380e+01 | 0.0000 | 200.0 | 5.24% | 124.5 | 3.10% | motif file (matrix) |
pdf |
| 11 |  | E-box/Arabidopsis-Promoters/Homer | 1e-5 | -1.335e+01 | 0.0001 | 69.0 | 1.81% | 26.8 | 0.67% | motif file (matrix) |
pdf |
| 12 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-5 | -1.307e+01 | 0.0001 | 268.0 | 7.02% | 184.2 | 4.58% | motif file (matrix) |
pdf |
| 13 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-5 | -1.303e+01 | 0.0001 | 62.0 | 1.62% | 22.9 | 0.57% | motif file (matrix) |
pdf |
| 14 |  | CLOCK(bHLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-5 | -1.195e+01 | 0.0002 | 83.0 | 2.17% | 38.4 | 0.96% | motif file (matrix) |
pdf |
| 15 |  | BMAL1(bHLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-4 | -1.045e+01 | 0.0008 | 319.0 | 8.36% | 241.2 | 6.00% | motif file (matrix) |
pdf |
| 16 |  | ZNF143|STAF(Zf)/CUTLL-ZNF143-ChIP-Seq(GSE29600)/Homer | 1e-4 | -1.032e+01 | 0.0008 | 57.0 | 1.49% | 23.0 | 0.57% | motif file (matrix) |
pdf |
| 17 |  | STAT4(Stat)/CD4-Stat4-ChIP-Seq(GSE22104)/Homer | 1e-4 | -1.031e+01 | 0.0008 | 140.0 | 3.67% | 86.7 | 2.16% | motif file (matrix) |
pdf |
| 18 |  | Pho2(bHLH)/Yeast-Pho2-ChIP-Seq(GSE29506)/Homer | 1e-4 | -1.023e+01 | 0.0008 | 46.0 | 1.21% | 16.7 | 0.42% | motif file (matrix) |
pdf |
| 19 |  | Usf2(bHLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-4 | -1.003e+01 | 0.0009 | 72.0 | 1.89% | 34.3 | 0.85% | motif file (matrix) |
pdf |
| 20 |  | Bapx1(Homeobox)/VertebralCol-Bapx1-ChIP-Seq(GSE36672)/Homer | 1e-4 | -9.838e+00 | 0.0010 | 477.0 | 12.50% | 391.0 | 9.72% | motif file (matrix) |
pdf |
| 21 |  | PIF4(bHLH)/Seedling-PIF4-ChIP-Seq(GSE35315)/Homer | 1e-4 | -9.784e+00 | 0.0010 | 211.0 | 5.53% | 148.5 | 3.69% | motif file (matrix) |
pdf |
| 22 |  | GAGA-repeat/SacCer-Promoters/Homer | 1e-4 | -9.376e+00 | 0.0015 | 804.0 | 21.06% | 711.2 | 17.68% | motif file (matrix) |
pdf |
| 23 |  | Unknown6/Drosophila-Promoters/Homer | 1e-4 | -9.375e+00 | 0.0015 | 243.0 | 6.37% | 178.1 | 4.43% | motif file (matrix) |
pdf |
| 24 |  | bHLHE40(bHLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.747e+00 | 0.0026 | 41.0 | 1.07% | 15.7 | 0.39% | motif file (matrix) |
pdf |
| 25 |  | STAT5(Stat)/mCD4+-Stat5-ChIP-Seq(GSE12346)/Homer | 1e-3 | -8.674e+00 | 0.0026 | 57.0 | 1.49% | 26.9 | 0.67% | motif file (matrix) |
pdf |
| 26 |  | c-Myc(bHLH)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-3 | -8.467e+00 | 0.0031 | 78.0 | 2.04% | 42.3 | 1.05% | motif file (matrix) |
pdf |
| 27 |  | Gfi1b(Zf)/HPC7-Gfi1b-ChIP-Seq(GSE22178)/Homer | 1e-3 | -8.153e+00 | 0.0041 | 136.0 | 3.56% | 90.1 | 2.24% | motif file (matrix) |
pdf |
| 28 |  | Pho4(bHLH)/Yeast-Pho4-ChIP-Seq(GSE29506)/Homer | 1e-3 | -7.939e+00 | 0.0049 | 36.0 | 0.94% | 13.1 | 0.33% | motif file (matrix) |
pdf |
| 29 |  | Cdx2(Homeobox)/mES-Cdx2-ChIP-Seq(GSE14586)/Homer | 1e-3 | -7.929e+00 | 0.0049 | 196.0 | 5.13% | 143.7 | 3.57% | motif file (matrix) |
pdf |
| 30 |  | Unknown4/Drosophila-Promoters/Homer | 1e-3 | -7.909e+00 | 0.0049 | 42.0 | 1.10% | 17.8 | 0.44% | motif file (matrix) |
pdf |
| 31 |  | USF1(bHLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-3 | -7.803e+00 | 0.0051 | 68.0 | 1.78% | 36.4 | 0.91% | motif file (matrix) |
pdf |
| 32 |  | Brn1(POU,Homeobox)/NPC-Brn1-ChIP-Seq(GSE35496)/Homer | 1e-3 | -7.747e+00 | 0.0052 | 82.0 | 2.15% | 47.4 | 1.18% | motif file (matrix) |
pdf |
| 33 |  | IBL1(bHLH)/Seedling-IBL1-ChIP-Seq(GSE51120)/Homer | 1e-3 | -7.596e+00 | 0.0059 | 221.0 | 5.79% | 167.2 | 4.16% | motif file (matrix) |
pdf |
| 34 |  | KLF14(Zf)/HEK293-KLF14.GFP-ChIP-Seq(GSE58341)/Homer | 1e-3 | -7.159e+00 | 0.0089 | 203.0 | 5.32% | 153.8 | 3.82% | motif file (matrix) |
pdf |
| 35 |  | Max(bHLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.016e+00 | 0.0099 | 99.0 | 2.59% | 63.8 | 1.59% | motif file (matrix) |
pdf |
| 36 |  | SPCH(bHLH)/Seedling-SPCH-ChIP-Seq(GSE57497)/Homer | 1e-2 | -6.846e+00 | 0.0114 | 161.0 | 4.22% | 117.3 | 2.92% | motif file (matrix) |
pdf |
| 37 |  | Sox10(HMG)/SciaticNerve-Sox3-ChIP-Seq(GSE35132)/Homer | 1e-2 | -6.776e+00 | 0.0119 | 250.0 | 6.55% | 198.7 | 4.94% | motif file (matrix) |
pdf |
| 38 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-2 | -6.771e+00 | 0.0119 | 214.0 | 5.61% | 165.5 | 4.12% | motif file (matrix) |
pdf |
| 39 |  | Cbf1(bHLH)/Yeast-Cbf1-ChIP-Seq(GSE29506)/Homer | 1e-2 | -6.700e+00 | 0.0122 | 36.0 | 0.94% | 15.4 | 0.38% | motif file (matrix) |
pdf |
| 40 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-2 | -6.542e+00 | 0.0139 | 85.0 | 2.23% | 53.1 | 1.32% | motif file (matrix) |
pdf |
| 41 |  | Oct6(POU,Homeobox)/NPC-Oct6-ChIP-Seq(GSE35496)/Homer | 1e-2 | -6.529e+00 | 0.0139 | 91.0 | 2.38% | 58.7 | 1.46% | motif file (matrix) |
pdf |
| 42 |  | KLF5(Zf)/LoVo-KLF5-ChIP-Seq(GSE49402)/Homer | 1e-2 | -6.493e+00 | 0.0139 | 167.0 | 4.38% | 124.3 | 3.09% | motif file (matrix) |
pdf |
| 43 |  | THRa(NR)/C17.2-THRa-ChIP-Seq(GSE38347)/Homer | 1e-2 | -6.422e+00 | 0.0146 | 71.0 | 1.86% | 42.3 | 1.05% | motif file (matrix) |
pdf |
| 44 |  | HIF-1b(HLH)/T47D-HIF1b-ChIP-Seq(GSE59937)/Homer | 1e-2 | -6.209e+00 | 0.0177 | 143.0 | 3.75% | 104.3 | 2.59% | motif file (matrix) |
pdf |
| 45 |  | Mef2c(MADS)/GM12878-Mef2c-ChIP-Seq(GSE32465)/Homer | 1e-2 | -6.017e+00 | 0.0210 | 142.0 | 3.72% | 104.9 | 2.61% | motif file (matrix) |
pdf |
| 46 |  | n-Myc(bHLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.997e+00 | 0.0210 | 90.0 | 2.36% | 59.1 | 1.47% | motif file (matrix) |
pdf |
| 47 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan_et_al.)/Homer | 1e-2 | -5.931e+00 | 0.0219 | 287.0 | 7.52% | 238.4 | 5.93% | motif file (matrix) |
pdf |
| 48 |  | Klf9(Zf)/GBM-Klf9-ChIP-Seq(GSE62211)/Homer | 1e-2 | -5.929e+00 | 0.0219 | 34.0 | 0.89% | 15.6 | 0.39% | motif file (matrix) |
pdf |
| 49 |  | FOXA1:AR(Forkhead,NR)/LNCAP-AR-ChIP-Seq(GSE27824)/Homer | 1e-2 | -5.919e+00 | 0.0219 | 15.0 | 0.39% | 3.1 | 0.08% | motif file (matrix) |
pdf |
| 50 |  | PIF5ox(bHLH)/Arabidopsis-PIF5ox-ChIP-Seq(GSE35062)/Homer | 1e-2 | -5.901e+00 | 0.0219 | 147.0 | 3.85% | 109.9 | 2.73% | motif file (matrix) |
pdf |
| 51 | 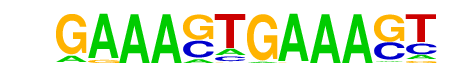 | IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer | 1e-2 | -5.437e+00 | 0.0330 | 24.0 | 0.63% | 9.1 | 0.23% | motif file (matrix) |
pdf |
| 52 |  | Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer | 1e-2 | -5.402e+00 | 0.0335 | 200.0 | 5.24% | 160.5 | 3.99% | motif file (matrix) |
pdf |
| 53 |  | MITF(bHLH)/MastCells-MITF-ChIP-Seq(GSE48085)/Homer | 1e-2 | -5.259e+00 | 0.0380 | 120.0 | 3.14% | 88.3 | 2.19% | motif file (matrix) |
pdf |
| 54 |  | Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer | 1e-2 | -4.977e+00 | 0.0494 | 415.0 | 10.87% | 369.8 | 9.20% | motif file (matrix) |
pdf |
| 55 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-2 | -4.886e+00 | 0.0531 | 27.0 | 0.71% | 13.0 | 0.32% | motif file (matrix) |
pdf |
| 56 |  | Oct4(POU,Homeobox)/mES-Oct4-ChIP-Seq(GSE11431)/Homer | 1e-2 | -4.734e+00 | 0.0608 | 106.0 | 2.78% | 78.8 | 1.96% | motif file (matrix) |
pdf |
| 57 |  | LXRE(NR),DR4/RAW-LXRb.biotin-ChIP-Seq(GSE21512)/Homer | 1e-2 | -4.713e+00 | 0.0610 | 32.0 | 0.84% | 16.2 | 0.40% | motif file (matrix) |
pdf |
| 58 |  | Reverb(NR),DR2/RAW-Reverba.biotin-ChIP-Seq(GSE45914)/Homer | 1e-2 | -4.660e+00 | 0.0631 | 25.0 | 0.65% | 11.8 | 0.29% | motif file (matrix) |
pdf |



















































































































