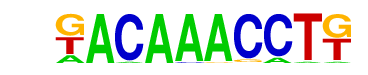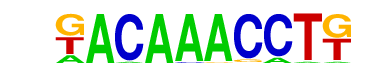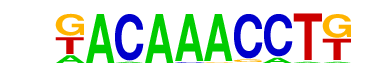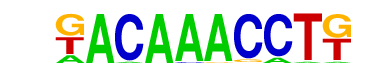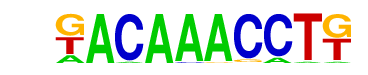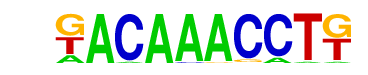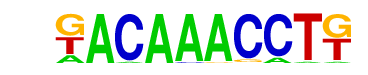| p-value: | 1e-7 |
| log p-value: | -1.722e+01 |
| Information Content per bp: | 1.829 |
| Number of Target Sequences with motif | 31.0 |
| Percentage of Target Sequences with motif | 2.30% |
| Number of Background Sequences with motif | 2.1 |
| Percentage of Background Sequences with motif | 0.15% |
| Average Position of motif in Targets | 35.7 +/- 18.1bp |
| Average Position of motif in Background | 45.0 +/- 0.0bp |
| Strand Bias (log2 ratio + to - strand density) | -0.9 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
ID1(C2H2(Zn))/Zea mays/AthaMap
| Match Rank: | 1 |
| Score: | 0.66
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----KACAAACCTK
AAAACGACAAA---- |
|


|
|
ZEB1/MA0103.2/Jaspar
| Match Rank: | 2 |
| Score: | 0.64
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | KACAAACCTK
-CCTCACCTG |
|

|
|
SD0001.1_at_AC_acceptor/Jaspar
| Match Rank: | 3 |
| Score: | 0.64
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -KACAAACCTK
NNACTTACCTN |
|


|
|
MF0011.1_HMG_class/Jaspar
| Match Rank: | 4 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | KACAAACCTK
AACAAT---- |
|
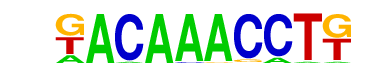

|
|
P(MYB)/Zea mays/AthaMap
| Match Rank: | 5 |
| Score: | 0.64
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | KACAAACCTK
-ACCTACCCG |
|
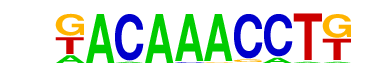

|
|
Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer
| Match Rank: | 6 |
| Score: | 0.63
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --KACAAACCTK
RNAACAATGG-- |
|


|
|
SOX10/MA0442.1/Jaspar
| Match Rank: | 7 |
| Score: | 0.63
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | KACAAACCTK
-ACAAAG--- |
|
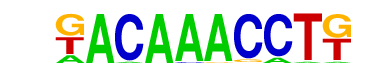

|
|
Sox10(HMG)/SciaticNerve-Sox3-ChIP-Seq(GSE35132)/Homer
| Match Rank: | 8 |
| Score: | 0.62
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --KACAAACCTK
VRRACAAWGG-- |
|


|
|
POL007.1_BREd/Jaspar
| Match Rank: | 9 |
| Score: | 0.62
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | KACAAACCTK
NANANAC--- |
|
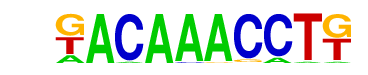

|
|
TBX15/MA0803.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.62
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | KACAAACCTK
-TCACACCT- |
|
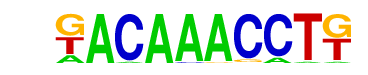

|
|