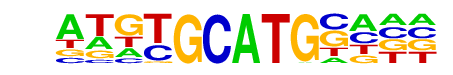
| p-value: | 1e-9 |
| log p-value: | -2.277e+01 |
| Information Content per bp: | 1.480 |
| Number of Target Sequences with motif | 68.0 |
| Percentage of Target Sequences with motif | 9.16% |
| Number of Background Sequences with motif | 11.9 |
| Percentage of Background Sequences with motif | 1.78% |
| Average Position of motif in Targets | 34.3 +/- 17.9bp |
| Average Position of motif in Background | 42.4 +/- 25.4bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.03 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
AtLEC2(ABI3/VP1)/Arabidopsis thaliana/AthaMap
| Match Rank: | 1 |
| Score: | 0.66
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GGCATGARCC
TTTGCATGGA-- |
|


|
|
RBFox2(?)/Heart-RBFox2-CLIP-Seq(GSE57926)/Homer
| Match Rank: | 2 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GGCATGARCC
TGCATGCA-- |
|


|
|
LEC2/MA0581.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.65
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GGCATGARCC
ATGTGCATGNN-- |
|

|
|
hkb/MA0450.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.64
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GGCATGARCC
GGGGCGTGA--- |
|


|
|
ZNF467(Zf)/HEK293-ZNF467.GFP-ChIP-Seq(GSE58341)/Homer
| Match Rank: | 5 |
| Score: | 0.64
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GGCATGARCC
TGGGGAAGGGCM |
|


|
|
ttk/dmmpmm(Papatsenko)/fly
| Match Rank: | 6 |
| Score: | 0.63
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GGCATGARCC
GCCAGGACC- |
|


|
|
TEC1/TEC1_YPD/[](Harbison)/Yeast
| Match Rank: | 7 |
| Score: | 0.63
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GGCATGARCC
AGGAATG---- |
|


|
|
E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer
| Match Rank: | 8 |
| Score: | 0.62
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GGCATGARCC
CWGGCGGGAA-- |
|


|
|
ABI3/MA0564.1/Jaspar
| Match Rank: | 9 |
| Score: | 0.62
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GGCATGARCC
CTGCATGCA-- |
|


|
|
hkb/dmmpmm(Noyes)/fly
| Match Rank: | 10 |
| Score: | 0.62
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GGCATGARCC
GGGGGCGTGANG- |
|


|
|





















