| p-value: | 1e-11 |
| log p-value: | -2.748e+01 |
| Information Content per bp: | 1.675 |
| Number of Target Sequences with motif | 133.0 |
| Percentage of Target Sequences with motif | 8.41% |
| Number of Background Sequences with motif | 45.1 |
| Percentage of Background Sequences with motif | 2.78% |
| Average Position of motif in Targets | 35.7 +/- 17.9bp |
| Average Position of motif in Background | 39.8 +/- 23.7bp |
| Strand Bias (log2 ratio + to - strand density) | 0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.02 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
RPN4/MA0373.1/Jaspar
| Match Rank: | 1 |
| Score: | 0.72
| | Offset: | 6
| | Orientation: | forward strand |
| Alignment: | DGGCDKGGTGGC-
------GGTGGCG |
|


|
|
RPN4/RPN4_H2O2Lo/[](Harbison)/Yeast
| Match Rank: | 2 |
| Score: | 0.66
| | Offset: | 6
| | Orientation: | reverse strand |
| Alignment: | DGGCDKGGTGGC---
------GGTGGCAAA |
|


|
|
MET32/MA0334.1/Jaspar
| Match Rank: | 3 |
| Score: | 0.66
| | Offset: | 6
| | Orientation: | reverse strand |
| Alignment: | DGGCDKGGTGGC-
------TGTGGCG |
|


|
|
MET31/MA0333.1/Jaspar
| Match Rank: | 4 |
| Score: | 0.66
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | DGGCDKGGTGGC-
----AGTGTGGCG |
|


|
|
RPN4(MacIsaac)/Yeast
| Match Rank: | 5 |
| Score: | 0.65
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | DGGCDKGGTGGC----
-----CGGTGGCAAAA |
|


|
|
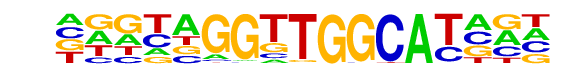
PB0029.1_Hic1_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.65
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | DGGCDKGGTGGC-----
-NGTAGGTTGGCATNNN |
|

|
|
POL006.1_BREu/Jaspar
| Match Rank: | 7 |
| Score: | 0.63
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | DGGCDKGGTGGC
-GGCGCGCT--- |
|


|
|
PB0039.1_Klf7_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.63
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----DGGCDKGGTGGC
NNAGGGGCGGGGTNNA |
|


|
|
AtMYB84(MYB)/Arabidopsis thaliana/AthaMap
| Match Rank: | 9 |
| Score: | 0.63
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --DGGCDKGGTGGC
GGGGGGTAGGTGG- |
|


|
|
RAX3/MA0576.1/Jaspar
| Match Rank: | 10 |
| Score: | 0.63
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --DGGCDKGGTGGC
GGGGGGTAGGTGG- |
|


|
|





















