| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | PHA-4(Forkhead)/cElegans-Embryos-PHA4-ChIP-Seq(modEncode)/Homer | 1e-5 | -1.328e+01 | 0.0007 | 272.0 | 17.19% | 185.6 | 11.44% | motif file (matrix) |
pdf |
| 2 |  | PABPC1(?)/MEL-PABC1-CLIP-Seq(GSE69755)/Homer | 1e-5 | -1.306e+01 | 0.0007 | 249.0 | 15.74% | 166.5 | 10.26% | motif file (matrix) |
pdf |
| 3 |  | FOXM1(Forkhead)/MCF7-FOXM1-ChIP-Seq(GSE72977)/Homer | 1e-5 | -1.211e+01 | 0.0007 | 89.0 | 5.63% | 41.1 | 2.53% | motif file (matrix) |
pdf |
| 4 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-5 | -1.190e+01 | 0.0007 | 94.0 | 5.94% | 45.6 | 2.81% | motif file (matrix) |
pdf |
| 5 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-5 | -1.170e+01 | 0.0007 | 107.0 | 6.76% | 55.6 | 3.43% | motif file (matrix) |
pdf |
| 6 |  | ZNF416(Zf)/HEK293-ZNF416.GFP-ChIP-Seq(GSE58341)/Homer | 1e-4 | -1.115e+01 | 0.0009 | 121.0 | 7.65% | 67.4 | 4.15% | motif file (matrix) |
pdf |
| 7 |  | Pho2(bHLH)/Yeast-Pho2-ChIP-Seq(GSE29506)/Homer | 1e-4 | -1.095e+01 | 0.0010 | 24.0 | 1.52% | 3.9 | 0.24% | motif file (matrix) |
pdf |
| 8 |  | Srebp2(bHLH)/HepG2-Srebp2-ChIP-Seq(GSE31477)/Homer | 1e-4 | -9.774e+00 | 0.0028 | 17.0 | 1.07% | 1.6 | 0.10% | motif file (matrix) |
pdf |
| 9 |  | FoxL2(Forkhead)/Ovary-FoxL2-ChIP-Seq(GSE60858)/Homer | 1e-3 | -8.489e+00 | 0.0088 | 86.0 | 5.44% | 47.1 | 2.91% | motif file (matrix) |
pdf |
| 10 |  | Bapx1(Homeobox)/VertebralCol-Bapx1-ChIP-Seq(GSE36672)/Homer | 1e-3 | -8.476e+00 | 0.0088 | 190.0 | 12.01% | 133.3 | 8.22% | motif file (matrix) |
pdf |
| 11 |  | GSC(Homeobox)/FrogEmbryos-GSC-ChIP-Seq(DRA000576)/Homer | 1e-3 | -8.065e+00 | 0.0111 | 188.0 | 11.88% | 133.9 | 8.25% | motif file (matrix) |
pdf |
| 12 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-3 | -8.035e+00 | 0.0111 | 25.0 | 1.58% | 6.4 | 0.40% | motif file (matrix) |
pdf |
| 13 |  | Pho4(bHLH)/Yeast-Pho4-ChIP-Seq(GSE29506)/Homer | 1e-3 | -8.002e+00 | 0.0111 | 19.0 | 1.20% | 3.2 | 0.20% | motif file (matrix) |
pdf |
| 14 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-3 | -7.170e+00 | 0.0213 | 13.0 | 0.82% | 1.9 | 0.12% | motif file (matrix) |
pdf |
| 15 |  | LHY(Myb)/Seedling-LHY-ChIP-Seq(GSE52175)/Homer | 1e-3 | -7.141e+00 | 0.0213 | 123.0 | 7.77% | 81.6 | 5.03% | motif file (matrix) |
pdf |
| 16 |  | Etv2(ETS)/ES-ER71-ChIP-Seq(GSE59402)/Homer(0.967) | 1e-3 | -7.076e+00 | 0.0213 | 45.0 | 2.84% | 21.0 | 1.29% | motif file (matrix) |
pdf |
| 17 |  | Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer | 1e-3 | -6.931e+00 | 0.0222 | 164.0 | 10.37% | 117.2 | 7.22% | motif file (matrix) |
pdf |
| 18 |  | Unknown4/Arabidopsis-Promoters/Homer | 1e-2 | -6.885e+00 | 0.0222 | 74.0 | 4.68% | 42.7 | 2.63% | motif file (matrix) |
pdf |
| 19 |  | CLOCK(bHLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-2 | -6.825e+00 | 0.0222 | 34.0 | 2.15% | 13.7 | 0.84% | motif file (matrix) |
pdf |
| 20 |  | SeqBias: TA-repeat | 1e-2 | -6.797e+00 | 0.0222 | 436.0 | 27.56% | 370.0 | 22.81% | motif file (matrix) |
pdf |
| 21 |  | GAGA-repeat/SacCer-Promoters/Homer | 1e-2 | -6.785e+00 | 0.0222 | 332.0 | 20.99% | 271.1 | 16.71% | motif file (matrix) |
pdf |
| 22 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-2 | -6.362e+00 | 0.0304 | 43.0 | 2.72% | 20.2 | 1.25% | motif file (matrix) |
pdf |
| 23 |  | Mef2c(MADS)/GM12878-Mef2c-ChIP-Seq(GSE32465)/Homer | 1e-2 | -6.128e+00 | 0.0352 | 70.0 | 4.42% | 41.5 | 2.56% | motif file (matrix) |
pdf |
| 24 | 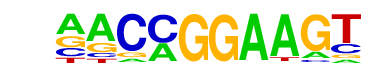 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-2 | -6.128e+00 | 0.0352 | 70.0 | 4.42% | 41.4 | 2.56% | motif file (matrix) |
pdf |
| 25 |  | Unknown2/Drosophila-Promoters/Homer | 1e-2 | -5.882e+00 | 0.0432 | 96.0 | 6.07% | 63.2 | 3.89% | motif file (matrix) |
pdf |
| 26 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-2 | -5.735e+00 | 0.0481 | 37.0 | 2.34% | 17.1 | 1.05% | motif file (matrix) |
pdf |
| 27 |  | Srebp1a(bHLH)/HepG2-Srebp1a-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.581e+00 | 0.0540 | 20.0 | 1.26% | 6.0 | 0.37% | motif file (matrix) |
pdf |
| 28 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-2 | -5.470e+00 | 0.0582 | 32.0 | 2.02% | 14.9 | 0.92% | motif file (matrix) |
pdf |
| 29 |  | USF1(bHLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.470e+00 | 0.0582 | 23.0 | 1.45% | 8.9 | 0.55% | motif file (matrix) |
pdf |
| 30 |  | bHLHE40(bHLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.436e+00 | 0.0582 | 18.0 | 1.14% | 5.7 | 0.35% | motif file (matrix) |
pdf |
| 31 |  | E-box/Arabidopsis-Promoters/Homer | 1e-2 | -5.234e+00 | 0.0666 | 24.0 | 1.52% | 9.1 | 0.56% | motif file (matrix) |
pdf |
| 32 |  | Sox4(HMG)/proB-Sox4-ChIP-Seq(GSE50066)/Homer | 1e-2 | -5.047e+00 | 0.0777 | 47.0 | 2.97% | 26.0 | 1.60% | motif file (matrix) |
pdf |
| 33 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-2 | -4.962e+00 | 0.0821 | 48.0 | 3.03% | 28.0 | 1.72% | motif file (matrix) |
pdf |
| 34 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-2 | -4.957e+00 | 0.0821 | 80.0 | 5.06% | 53.1 | 3.27% | motif file (matrix) |
pdf |
| 35 |  | Cdx2(Homeobox)/mES-Cdx2-ChIP-Seq(GSE14586)/Homer | 1e-2 | -4.793e+00 | 0.0917 | 84.0 | 5.31% | 58.0 | 3.57% | motif file (matrix) |
pdf |
| 36 |  | Sox15(HMG)/CPA-Sox15-ChIP-Seq(GSE62909)/Homer | 1e-2 | -4.774e+00 | 0.0917 | 78.0 | 4.93% | 52.1 | 3.21% | motif file (matrix) |
pdf |
| 37 |  | Mef2d(MADS)/Retina-Mef2d-ChIP-Seq(GSE61391)/Homer | 1e-2 | -4.633e+00 | 0.1017 | 24.0 | 1.52% | 10.6 | 0.65% | motif file (matrix) |
pdf |









































































