| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-88 | -2.037e+02 | 0.0000 | 382.0 | 78.93% | 51.1 | 13.32% | motif file (matrix) |
pdf |
| 2 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-62 | -1.450e+02 | 0.0000 | 303.0 | 62.60% | 35.6 | 9.26% | motif file (matrix) |
pdf |
| 3 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-58 | -1.348e+02 | 0.0000 | 267.0 | 55.17% | 23.5 | 6.13% | motif file (matrix) |
pdf |
| 4 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-55 | -1.282e+02 | 0.0000 | 250.0 | 51.65% | 19.8 | 5.15% | motif file (matrix) |
pdf |
| 5 |  | ELT-3(Gata)/cElegans-L1-ELT3-ChIP-Seq(modEncode)/Homer | 1e-15 | -3.546e+01 | 0.0000 | 97.0 | 20.04% | 11.9 | 3.09% | motif file (matrix) |
pdf |
| 6 |  | GATA:SCL(Zf,bHLH)/Ter119-SCL-ChIP-Seq(GSE18720)/Homer | 1e-9 | -2.109e+01 | 0.0000 | 44.0 | 9.09% | 2.9 | 0.76% | motif file (matrix) |
pdf |
| 7 |  | PQM-1(?)/cElegans-L3-ChIP-Seq(modEncode)/Homer | 1e-8 | -2.046e+01 | 0.0000 | 76.0 | 15.70% | 14.3 | 3.71% | motif file (matrix) |
pdf |
| 8 |  | CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer | 1e-6 | -1.436e+01 | 0.0000 | 138.0 | 28.51% | 56.2 | 14.62% | motif file (matrix) |
pdf |
| 9 |  | BMYB(HTH)/Hela-BMYB-ChIP-Seq(GSE27030)/Homer | 1e-5 | -1.313e+01 | 0.0001 | 66.0 | 13.64% | 17.4 | 4.53% | motif file (matrix) |
pdf |
| 10 |  | Unknown5/Drosophila-Promoters/Homer | 1e-5 | -1.249e+01 | 0.0001 | 54.0 | 11.16% | 12.8 | 3.35% | motif file (matrix) |
pdf |
| 11 |  | AMYB(HTH)/Testes-AMYB-ChIP-Seq(GSE44588)/Homer | 1e-5 | -1.153e+01 | 0.0003 | 60.0 | 12.40% | 16.4 | 4.27% | motif file (matrix) |
pdf |
| 12 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-4 | -1.119e+01 | 0.0004 | 65.0 | 13.43% | 19.8 | 5.17% | motif file (matrix) |
pdf |
| 13 | 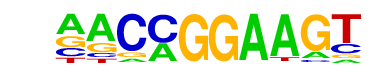 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-4 | -1.044e+01 | 0.0009 | 63.0 | 13.02% | 19.4 | 5.04% | motif file (matrix) |
pdf |
| 14 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-4 | -9.990e+00 | 0.0013 | 27.0 | 5.58% | 3.7 | 0.97% | motif file (matrix) |
pdf |
| 15 |  | HLH-1(bHLH)/cElegans-Embryo-HLH1-ChIP-Seq(modEncode)/Homer | 1e-3 | -8.928e+00 | 0.0034 | 30.0 | 6.20% | 5.5 | 1.42% | motif file (matrix) |
pdf |
| 16 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.900e+00 | 0.0034 | 22.0 | 4.55% | 2.4 | 0.62% | motif file (matrix) |
pdf |
| 17 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-3 | -8.485e+00 | 0.0047 | 38.0 | 7.85% | 9.5 | 2.46% | motif file (matrix) |
pdf |
| 18 |  | SeqBias: GA-repeat | 1e-3 | -7.826e+00 | 0.0086 | 225.0 | 46.49% | 134.6 | 35.05% | motif file (matrix) |
pdf |
| 19 |  | MyoD(bHLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-3 | -7.681e+00 | 0.0094 | 36.0 | 7.44% | 9.5 | 2.47% | motif file (matrix) |
pdf |
| 20 |  | STAT5(Stat)/mCD4+-Stat5-ChIP-Seq(GSE12346)/Homer | 1e-3 | -7.651e+00 | 0.0094 | 13.0 | 2.69% | 0.3 | 0.07% | motif file (matrix) |
pdf |
| 21 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.651e+00 | 0.0094 | 13.0 | 2.69% | 0.9 | 0.24% | motif file (matrix) |
pdf |
| 22 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-3 | -7.449e+00 | 0.0098 | 29.0 | 5.99% | 6.5 | 1.69% | motif file (matrix) |
pdf |
| 23 |  | KLF14(Zf)/HEK293-KLF14.GFP-ChIP-Seq(GSE58341)/Homer | 1e-3 | -7.449e+00 | 0.0098 | 29.0 | 5.99% | 6.4 | 1.66% | motif file (matrix) |
pdf |
| 24 |  | ZNF711(Zf)/SHSY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer | 1e-3 | -7.239e+00 | 0.0116 | 52.0 | 10.74% | 19.0 | 4.94% | motif file (matrix) |
pdf |
| 25 |  | STAT1(Stat)/HelaS3-STAT1-ChIP-Seq(GSE12782)/Homer | 1e-3 | -7.056e+00 | 0.0133 | 12.0 | 2.48% | 0.2 | 0.05% | motif file (matrix) |
pdf |
| 26 | 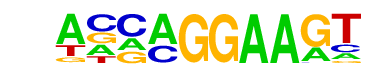 | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-2 | -6.825e+00 | 0.0162 | 56.0 | 11.57% | 21.8 | 5.67% | motif file (matrix) |
pdf |
| 27 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -6.620e+00 | 0.0191 | 41.0 | 8.47% | 13.9 | 3.61% | motif file (matrix) |
pdf |
| 28 |  | FoxL2(Forkhead)/Ovary-FoxL2-ChIP-Seq(GSE60858)/Homer | 1e-2 | -6.606e+00 | 0.0191 | 27.0 | 5.58% | 6.4 | 1.67% | motif file (matrix) |
pdf |
| 29 |  | Pdx1(Homeobox)/Islet-Pdx1-ChIP-Seq(SRA008281)/Homer | 1e-2 | -6.591e+00 | 0.0191 | 50.0 | 10.33% | 18.6 | 4.83% | motif file (matrix) |
pdf |
| 30 |  | Fra1(bZIP)/BT549-Fra1-ChIP-Seq(GSE46166)/Homer | 1e-2 | -6.565e+00 | 0.0191 | 20.0 | 4.13% | 3.8 | 1.00% | motif file (matrix) |
pdf |
| 31 |  | BMAL1(bHLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-2 | -6.452e+00 | 0.0197 | 46.0 | 9.50% | 16.1 | 4.20% | motif file (matrix) |
pdf |
| 32 |  | LHY(Myb)/Seedling-LHY-ChIP-Seq(GSE52175)/Homer | 1e-2 | -6.276e+00 | 0.0227 | 40.0 | 8.26% | 13.0 | 3.39% | motif file (matrix) |
pdf |
| 33 |  | Fosl2(bZIP)/3T3L1-Fosl2-ChIP-Seq(GSE56872)/Homer | 1e-2 | -6.260e+00 | 0.0227 | 14.0 | 2.89% | 1.3 | 0.33% | motif file (matrix) |
pdf |
| 34 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-2 | -6.260e+00 | 0.0227 | 14.0 | 2.89% | 1.8 | 0.48% | motif file (matrix) |
pdf |
| 35 |  | Tcf12(bHLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.886e+00 | 0.0307 | 37.0 | 7.64% | 12.7 | 3.30% | motif file (matrix) |
pdf |
| 36 |  | Zfp809(Zf)/ES-Zfp809-ChIP-Seq(GSE70799)/Homer | 1e-2 | -5.871e+00 | 0.0307 | 10.0 | 2.07% | 0.0 | 0.00% | motif file (matrix) |
pdf |
| 37 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-2 | -5.765e+00 | 0.0328 | 27.0 | 5.58% | 7.8 | 2.03% | motif file (matrix) |
pdf |
| 38 |  | CLOCK(bHLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-2 | -5.638e+00 | 0.0363 | 18.0 | 3.72% | 3.8 | 0.98% | motif file (matrix) |
pdf |
| 39 |  | Otx2(Homeobox)/EpiLC-Otx2-ChIP-Seq(GSE56098)/Homer | 1e-2 | -5.351e+00 | 0.0471 | 39.0 | 8.06% | 14.7 | 3.82% | motif file (matrix) |
pdf |
| 40 |  | GATA3(Zf),DR8/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.280e+00 | 0.0493 | 9.0 | 1.86% | 0.8 | 0.21% | motif file (matrix) |
pdf |
| 41 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer | 1e-2 | -5.204e+00 | 0.0519 | 12.0 | 2.48% | 2.0 | 0.52% | motif file (matrix) |
pdf |
| 42 |  | GATA3(Zf),DR4/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -5.185e+00 | 0.0519 | 17.0 | 3.51% | 3.8 | 0.98% | motif file (matrix) |
pdf |
| 43 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer | 1e-2 | -5.003e+00 | 0.0605 | 23.0 | 4.75% | 6.9 | 1.80% | motif file (matrix) |
pdf |
| 44 |  | Usf2(bHLH)/C2C12-Usf2-ChIP-Seq(GSE36030)/Homer | 1e-2 | -4.740e+00 | 0.0768 | 16.0 | 3.31% | 3.1 | 0.80% | motif file (matrix) |
pdf |
| 45 |  | FOXM1(Forkhead)/MCF7-FOXM1-ChIP-Seq(GSE72977)/Homer | 1e-2 | -4.660e+00 | 0.0814 | 18.0 | 3.72% | 5.0 | 1.29% | motif file (matrix) |
pdf |
| 46 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-2 | -4.626e+00 | 0.0824 | 20.0 | 4.13% | 5.6 | 1.45% | motif file (matrix) |
pdf |
| 47 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-2 | -4.622e+00 | 0.0824 | 22.0 | 4.55% | 6.2 | 1.61% | motif file (matrix) |
pdf |
| 48 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-2 | -4.607e+00 | 0.0824 | 40.0 | 8.26% | 16.0 | 4.18% | motif file (matrix) |
pdf |































































































