| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | GATA3(Zf)/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-617 | -1.421e+03 | 0.0000 | 2315.0 | 77.17% | 230.4 | 9.21% | motif file (matrix) |
pdf |
| 2 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-503 | -1.159e+03 | 0.0000 | 1974.0 | 65.80% | 158.1 | 6.32% | motif file (matrix) |
pdf |
| 3 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-430 | -9.923e+02 | 0.0000 | 1694.0 | 56.47% | 104.1 | 4.16% | motif file (matrix) |
pdf |
| 4 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-402 | -9.274e+02 | 0.0000 | 1604.0 | 53.47% | 95.9 | 3.83% | motif file (matrix) |
pdf |
| 5 |  | ELT-3(Gata)/cElegans-L1-ELT3-ChIP-Seq(modEncode)/Homer | 1e-110 | -2.535e+02 | 0.0000 | 666.0 | 22.20% | 74.0 | 2.96% | motif file (matrix) |
pdf |
| 6 |  | GATA:SCL(Zf,bHLH)/Ter119-SCL-ChIP-Seq(GSE18720)/Homer | 1e-110 | -2.534e+02 | 0.0000 | 481.0 | 16.03% | 16.8 | 0.67% | motif file (matrix) |
pdf |
| 7 |  | PQM-1(?)/cElegans-L3-ChIP-Seq(modEncode)/Homer | 1e-105 | -2.434e+02 | 0.0000 | 625.0 | 20.83% | 65.3 | 2.61% | motif file (matrix) |
pdf |
| 8 |  | Unknown5/Drosophila-Promoters/Homer | 1e-65 | -1.509e+02 | 0.0000 | 482.0 | 16.07% | 72.7 | 2.90% | motif file (matrix) |
pdf |
| 9 |  | CRX(Homeobox)/Retina-Crx-ChIP-Seq(GSE20012)/Homer | 1e-26 | -6.162e+01 | 0.0000 | 816.0 | 27.20% | 381.1 | 15.23% | motif file (matrix) |
pdf |
| 10 |  | SCL(bHLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-24 | -5.646e+01 | 0.0000 | 1153.0 | 38.43% | 635.5 | 25.40% | motif file (matrix) |
pdf |
| 11 |  | GATA3(Zf),DR4/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-21 | -5.025e+01 | 0.0000 | 102.0 | 3.40% | 4.2 | 0.17% | motif file (matrix) |
pdf |
| 12 |  | SeqBias: polyA-repeat | 1e-21 | -4.861e+01 | 0.0000 | 2945.0 | 98.17% | 2327.2 | 93.03% | motif file (matrix) |
pdf |
| 13 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-21 | -4.842e+01 | 0.0000 | 219.0 | 7.30% | 49.8 | 1.99% | motif file (matrix) |
pdf |
| 14 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-20 | -4.835e+01 | 0.0000 | 221.0 | 7.37% | 50.4 | 2.01% | motif file (matrix) |
pdf |
| 15 |  | Tcf21(bHLH)/ArterySmoothMuscle-Tcf21-ChIP-Seq(GSE61369)/Homer | 1e-20 | -4.830e+01 | 0.0000 | 309.0 | 10.30% | 94.7 | 3.79% | motif file (matrix) |
pdf |
| 16 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-20 | -4.680e+01 | 0.0000 | 228.0 | 7.60% | 55.6 | 2.22% | motif file (matrix) |
pdf |
| 17 | 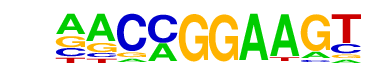 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-19 | -4.543e+01 | 0.0000 | 327.0 | 10.90% | 109.0 | 4.36% | motif file (matrix) |
pdf |
| 18 |  | Etv2(ETS)/ES-ER71-ChIP-Seq(GSE59402)/Homer(0.967) | 1e-19 | -4.438e+01 | 0.0000 | 218.0 | 7.27% | 54.0 | 2.16% | motif file (matrix) |
pdf |
| 19 |  | Fra1(bZIP)/BT549-Fra1-ChIP-Seq(GSE46166)/Homer | 1e-18 | -4.328e+01 | 0.0000 | 194.0 | 6.47% | 43.5 | 1.74% | motif file (matrix) |
pdf |
| 20 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-18 | -4.242e+01 | 0.0000 | 192.0 | 6.40% | 43.4 | 1.73% | motif file (matrix) |
pdf |
| 21 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-18 | -4.226e+01 | 0.0000 | 348.0 | 11.60% | 125.3 | 5.01% | motif file (matrix) |
pdf |
| 22 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-18 | -4.157e+01 | 0.0000 | 316.0 | 10.53% | 108.0 | 4.32% | motif file (matrix) |
pdf |
| 23 |  | Ap4(bHLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-17 | -4.131e+01 | 0.0000 | 376.0 | 12.53% | 143.9 | 5.75% | motif file (matrix) |
pdf |
| 24 |  | MyoD(bHLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-17 | -4.001e+01 | 0.0000 | 247.0 | 8.23% | 73.2 | 2.92% | motif file (matrix) |
pdf |
| 25 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer | 1e-17 | -3.987e+01 | 0.0000 | 186.0 | 6.20% | 43.8 | 1.75% | motif file (matrix) |
pdf |
| 26 | 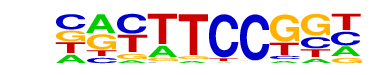 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-17 | -3.917e+01 | 0.0000 | 239.0 | 7.97% | 70.5 | 2.82% | motif file (matrix) |
pdf |
| 27 |  | Ascl1(bHLH)/NeuralTubes-Ascl1-ChIP-Seq(GSE55840)/Homer | 1e-16 | -3.836e+01 | 0.0000 | 385.0 | 12.83% | 154.8 | 6.19% | motif file (matrix) |
pdf |
| 28 |  | Tcf12(bHLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-16 | -3.823e+01 | 0.0000 | 292.0 | 9.73% | 100.8 | 4.03% | motif file (matrix) |
pdf |
| 29 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-16 | -3.706e+01 | 0.0000 | 202.0 | 6.73% | 54.5 | 2.18% | motif file (matrix) |
pdf |
| 30 |  | Fosl2(bZIP)/3T3L1-Fosl2-ChIP-Seq(GSE56872)/Homer | 1e-15 | -3.653e+01 | 0.0000 | 154.0 | 5.13% | 32.4 | 1.30% | motif file (matrix) |
pdf |
| 31 |  | Ptf1a(bHLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer | 1e-15 | -3.504e+01 | 0.0000 | 615.0 | 20.50% | 311.2 | 12.44% | motif file (matrix) |
pdf |
| 32 |  | HLH-1(bHLH)/cElegans-Embryo-HLH1-ChIP-Seq(modEncode)/Homer | 1e-14 | -3.344e+01 | 0.0000 | 214.0 | 7.13% | 66.0 | 2.64% | motif file (matrix) |
pdf |
| 33 |  | Olig2(bHLH)/Neuron-Olig2-ChIP-Seq(GSE30882)/Homer | 1e-14 | -3.261e+01 | 0.0000 | 451.0 | 15.03% | 208.9 | 8.35% | motif file (matrix) |
pdf |
| 34 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-13 | -3.195e+01 | 0.0000 | 234.0 | 7.80% | 78.4 | 3.13% | motif file (matrix) |
pdf |
| 35 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-13 | -3.141e+01 | 0.0000 | 197.0 | 6.57% | 59.8 | 2.39% | motif file (matrix) |
pdf |
| 36 | 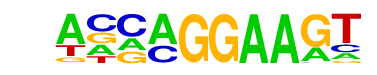 | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-13 | -3.080e+01 | 0.0000 | 285.0 | 9.50% | 109.6 | 4.38% | motif file (matrix) |
pdf |
| 37 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-13 | -3.062e+01 | 0.0000 | 221.0 | 7.37% | 73.8 | 2.95% | motif file (matrix) |
pdf |
| 38 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-13 | -3.034e+01 | 0.0000 | 124.0 | 4.13% | 25.8 | 1.03% | motif file (matrix) |
pdf |
| 39 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-12 | -2.964e+01 | 0.0000 | 229.0 | 7.63% | 79.5 | 3.18% | motif file (matrix) |
pdf |
| 40 |  | MyoG(bHLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-12 | -2.813e+01 | 0.0000 | 283.0 | 9.43% | 113.4 | 4.53% | motif file (matrix) |
pdf |
| 41 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-12 | -2.788e+01 | 0.0000 | 229.0 | 7.63% | 82.4 | 3.30% | motif file (matrix) |
pdf |
| 42 |  | HEB(bHLH)/mES-Heb-ChIP-Seq(GSE53233)/Homer | 1e-9 | -2.294e+01 | 0.0000 | 406.0 | 13.53% | 204.8 | 8.19% | motif file (matrix) |
pdf |
| 43 |  | BMYB(HTH)/Hela-BMYB-ChIP-Seq(GSE27030)/Homer | 1e-9 | -2.278e+01 | 0.0000 | 320.0 | 10.67% | 148.6 | 5.94% | motif file (matrix) |
pdf |
| 44 | 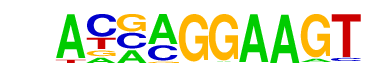 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-9 | -2.271e+01 | 0.0000 | 174.0 | 5.80% | 60.6 | 2.42% | motif file (matrix) |
pdf |
| 45 |  | SpiB(ETS)/OCILY3-SPIB-ChIP-Seq(GSE56857)/Homer | 1e-9 | -2.246e+01 | 0.0000 | 73.0 | 2.43% | 11.7 | 0.47% | motif file (matrix) |
pdf |
| 46 |  | Pdx1(Homeobox)/Islet-Pdx1-ChIP-Seq(SRA008281)/Homer | 1e-9 | -2.219e+01 | 0.0000 | 221.0 | 7.37% | 88.2 | 3.53% | motif file (matrix) |
pdf |
| 47 |  | Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer | 1e-9 | -2.194e+01 | 0.0000 | 456.0 | 15.20% | 241.4 | 9.65% | motif file (matrix) |
pdf |
| 48 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-8 | -2.067e+01 | 0.0000 | 157.0 | 5.23% | 54.7 | 2.19% | motif file (matrix) |
pdf |
| 49 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-8 | -2.038e+01 | 0.0000 | 134.0 | 4.47% | 42.5 | 1.70% | motif file (matrix) |
pdf |
| 50 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-8 | -2.012e+01 | 0.0000 | 192.0 | 6.40% | 75.8 | 3.03% | motif file (matrix) |
pdf |
| 51 | 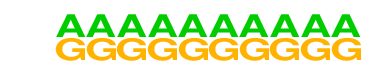 | SeqBias: G/A bias | 1e-8 | -1.992e+01 | 0.0000 | 2973.0 | 99.10% | 2424.3 | 96.92% | motif file (matrix) |
pdf |
| 52 |  | E2A(bHLH)/proBcell-E2A-ChIP-Seq(GSE21978)/Homer | 1e-8 | -1.965e+01 | 0.0000 | 318.0 | 10.60% | 155.9 | 6.23% | motif file (matrix) |
pdf |
| 53 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski_et_al.)/Homer | 1e-8 | -1.922e+01 | 0.0000 | 89.0 | 2.97% | 21.1 | 0.84% | motif file (matrix) |
pdf |
| 54 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-8 | -1.887e+01 | 0.0000 | 86.0 | 2.87% | 20.1 | 0.80% | motif file (matrix) |
pdf |
| 55 |  | ETS:E-box(ETS,bHLH)/HPC7-Scl-ChIP-Seq(GSE22178)/Homer | 1e-8 | -1.872e+01 | 0.0000 | 46.0 | 1.53% | 4.4 | 0.18% | motif file (matrix) |
pdf |
| 56 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-7 | -1.826e+01 | 0.0000 | 126.0 | 4.20% | 41.4 | 1.66% | motif file (matrix) |
pdf |
| 57 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-7 | -1.783e+01 | 0.0000 | 346.0 | 11.53% | 179.7 | 7.18% | motif file (matrix) |
pdf |
| 58 |  | AMYB(HTH)/Testes-AMYB-ChIP-Seq(GSE44588)/Homer | 1e-7 | -1.746e+01 | 0.0000 | 310.0 | 10.33% | 156.3 | 6.25% | motif file (matrix) |
pdf |
| 59 |  | Tal1 | 1e-7 | -1.738e+01 | 0.0000 | 335.0 | 11.17% | 173.7 | 6.94% | motif file (matrix) |
pdf |
| 60 |  | Otx2(Homeobox)/EpiLC-Otx2-ChIP-Seq(GSE56098)/Homer | 1e-7 | -1.660e+01 | 0.0000 | 188.0 | 6.27% | 80.7 | 3.23% | motif file (matrix) |
pdf |
| 61 |  | GSC(Homeobox)/FrogEmbryos-GSC-ChIP-Seq(DRA000576)/Homer | 1e-6 | -1.563e+01 | 0.0000 | 353.0 | 11.77% | 191.6 | 7.66% | motif file (matrix) |
pdf |
| 62 |  | SeqBias: GA-repeat | 1e-6 | -1.559e+01 | 0.0000 | 1195.0 | 39.83% | 829.7 | 33.17% | motif file (matrix) |
pdf |
| 63 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-6 | -1.523e+01 | 0.0000 | 85.0 | 2.83% | 24.3 | 0.97% | motif file (matrix) |
pdf |
| 64 |  | LIN-39(Homeobox)/cElegans.L3-LIN39-ChIP-Seq(modEncode)/Homer | 1e-6 | -1.460e+01 | 0.0000 | 198.0 | 6.60% | 91.8 | 3.67% | motif file (matrix) |
pdf |
| 65 |  | PIF5ox(bHLH)/Arabidopsis-PIF5ox-ChIP-Seq(GSE35062)/Homer | 1e-6 | -1.439e+01 | 0.0000 | 130.0 | 4.33% | 50.8 | 2.03% | motif file (matrix) |
pdf |
| 66 |  | ETS(ETS)/Promoter/Homer | 1e-6 | -1.398e+01 | 0.0000 | 59.0 | 1.97% | 13.9 | 0.56% | motif file (matrix) |
pdf |
| 67 |  | GAGA-repeat/SacCer-Promoters/Homer | 1e-5 | -1.348e+01 | 0.0000 | 772.0 | 25.73% | 509.3 | 20.36% | motif file (matrix) |
pdf |
| 68 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan_et_al.)/Homer | 1e-5 | -1.344e+01 | 0.0000 | 346.0 | 11.53% | 194.0 | 7.76% | motif file (matrix) |
pdf |
| 69 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.332e+01 | 0.0000 | 53.0 | 1.77% | 11.6 | 0.46% | motif file (matrix) |
pdf |
| 70 |  | ZBTB18(Zf)/HEK293-ZBTB18.GFP-ChIP-Seq(GSE58341)/Homer | 1e-5 | -1.292e+01 | 0.0000 | 125.0 | 4.17% | 50.2 | 2.01% | motif file (matrix) |
pdf |
| 71 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.268e+01 | 0.0000 | 25.0 | 0.83% | 1.2 | 0.05% | motif file (matrix) |
pdf |
| 72 |  | E-box/Drosophila-Promoters/Homer | 1e-5 | -1.255e+01 | 0.0000 | 96.0 | 3.20% | 34.8 | 1.39% | motif file (matrix) |
pdf |
| 73 |  | n-Myc(bHLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-5 | -1.156e+01 | 0.0001 | 71.0 | 2.37% | 22.4 | 0.90% | motif file (matrix) |
pdf |
| 74 | 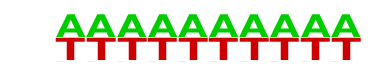 | SeqBias: A/T bias | 1e-4 | -1.147e+01 | 0.0001 | 2137.0 | 71.23% | 1647.9 | 65.88% | motif file (matrix) |
pdf |
| 75 |  | c-Myc(bHLH)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-4 | -1.072e+01 | 0.0001 | 51.0 | 1.70% | 14.0 | 0.56% | motif file (matrix) |
pdf |
| 76 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-4 | -1.061e+01 | 0.0001 | 81.0 | 2.70% | 29.9 | 1.20% | motif file (matrix) |
pdf |
| 77 |  | FOXM1(Forkhead)/MCF7-FOXM1-ChIP-Seq(GSE72977)/Homer | 1e-4 | -1.001e+01 | 0.0002 | 132.0 | 4.40% | 61.8 | 2.47% | motif file (matrix) |
pdf |
| 78 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-4 | -9.521e+00 | 0.0004 | 74.0 | 2.47% | 28.0 | 1.12% | motif file (matrix) |
pdf |
| 79 |  | Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer | 1e-3 | -9.202e+00 | 0.0005 | 264.0 | 8.80% | 153.2 | 6.12% | motif file (matrix) |
pdf |
| 80 |  | ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer | 1e-3 | -9.132e+00 | 0.0005 | 228.0 | 7.60% | 128.1 | 5.12% | motif file (matrix) |
pdf |
| 81 |  | Pho2(bHLH)/Yeast-Pho2-ChIP-Seq(GSE29506)/Homer | 1e-3 | -8.563e+00 | 0.0009 | 33.0 | 1.10% | 7.6 | 0.30% | motif file (matrix) |
pdf |
| 82 |  | Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.548e+00 | 0.0009 | 26.0 | 0.87% | 4.4 | 0.18% | motif file (matrix) |
pdf |
| 83 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-3 | -8.538e+00 | 0.0009 | 81.0 | 2.70% | 33.2 | 1.33% | motif file (matrix) |
pdf |
| 84 |  | Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.438e+00 | 0.0010 | 130.0 | 4.33% | 64.8 | 2.59% | motif file (matrix) |
pdf |
| 85 |  | ZNF189(Zf)/HEK293-ZNF189.GFP-ChIP-Seq(GSE58341)/Homer | 1e-3 | -8.282e+00 | 0.0012 | 106.0 | 3.53% | 49.5 | 1.98% | motif file (matrix) |
pdf |
| 86 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-3 | -8.031e+00 | 0.0015 | 167.0 | 5.57% | 90.8 | 3.63% | motif file (matrix) |
pdf |
| 87 |  | STAT1(Stat)/HelaS3-STAT1-ChIP-Seq(GSE12782)/Homer | 1e-3 | -8.026e+00 | 0.0015 | 38.0 | 1.27% | 11.0 | 0.44% | motif file (matrix) |
pdf |
| 88 |  | Nr5a2(NR)/Pancreas-LRH1-ChIP-Seq(GSE34295)/Homer | 1e-3 | -7.666e+00 | 0.0021 | 94.0 | 3.13% | 43.0 | 1.72% | motif file (matrix) |
pdf |
| 89 |  | Unknown-ESC-element(?)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-3 | -7.599e+00 | 0.0022 | 114.0 | 3.80% | 56.6 | 2.26% | motif file (matrix) |
pdf |
| 90 |  | ZNF711(Zf)/SHSY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer | 1e-3 | -7.513e+00 | 0.0023 | 273.0 | 9.10% | 167.9 | 6.71% | motif file (matrix) |
pdf |
| 91 | 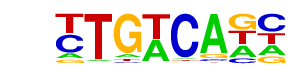 | Tgif1(Homeobox)/mES-Tgif1-ChIP-Seq(GSE55404)/Homer | 1e-3 | -7.402e+00 | 0.0026 | 502.0 | 16.73% | 339.1 | 13.56% | motif file (matrix) |
pdf |
| 92 |  | PIF4(bHLH)/Seedling-PIF4-ChIP-Seq(GSE35315)/Homer | 1e-3 | -7.178e+00 | 0.0032 | 145.0 | 4.83% | 78.1 | 3.12% | motif file (matrix) |
pdf |
| 93 |  | STAT5(Stat)/mCD4+-Stat5-ChIP-Seq(GSE12346)/Homer | 1e-3 | -7.153e+00 | 0.0032 | 47.0 | 1.57% | 16.4 | 0.66% | motif file (matrix) |
pdf |
| 94 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-3 | -7.153e+00 | 0.0032 | 47.0 | 1.57% | 16.6 | 0.66% | motif file (matrix) |
pdf |
| 95 |  | Smad2(MAD)/ES-SMAD2-ChIP-Seq(GSE29422)/Homer | 1e-3 | -7.067e+00 | 0.0035 | 259.0 | 8.63% | 159.3 | 6.37% | motif file (matrix) |
pdf |
| 96 |  | BMAL1(bHLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-2 | -6.888e+00 | 0.0041 | 226.0 | 7.53% | 136.1 | 5.44% | motif file (matrix) |
pdf |
| 97 |  | GATA3(Zf),DR8/iTreg-Gata3-ChIP-Seq(GSE20898)/Homer | 1e-2 | -6.867e+00 | 0.0042 | 33.0 | 1.10% | 9.2 | 0.37% | motif file (matrix) |
pdf |
| 98 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-2 | -6.830e+00 | 0.0043 | 89.0 | 2.97% | 42.5 | 1.70% | motif file (matrix) |
pdf |
| 99 |  | SeqBias: polyC-repeat | 1e-2 | -6.474e+00 | 0.0060 | 2697.0 | 89.90% | 2185.0 | 87.35% | motif file (matrix) |
pdf |
| 100 |  | Meis1(Homeobox)/MastCells-Meis1-ChIP-Seq(GSE48085)/Homer | 1e-2 | -6.358e+00 | 0.0067 | 265.0 | 8.83% | 167.7 | 6.70% | motif file (matrix) |
pdf |
| 101 |  | Initiator/Drosophila-Promoters/Homer | 1e-2 | -6.241e+00 | 0.0075 | 350.0 | 11.67% | 231.6 | 9.26% | motif file (matrix) |
pdf |
| 102 |  | Klf4(Zf)/mES-Klf4-ChIP-Seq(GSE11431)/Homer | 1e-2 | -6.180e+00 | 0.0079 | 44.0 | 1.47% | 16.7 | 0.67% | motif file (matrix) |
pdf |
| 103 |  | MITF(bHLH)/MastCells-MITF-ChIP-Seq(GSE48085)/Homer | 1e-2 | -6.089e+00 | 0.0085 | 132.0 | 4.40% | 73.5 | 2.94% | motif file (matrix) |
pdf |
| 104 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-2 | -5.971e+00 | 0.0095 | 160.0 | 5.33% | 93.1 | 3.72% | motif file (matrix) |
pdf |
| 105 |  | Egr2(Zf)/Thymocytes-Egr2-ChIP-Seq(GSE34254)/Homer | 1e-2 | -5.959e+00 | 0.0095 | 13.0 | 0.43% | 1.8 | 0.07% | motif file (matrix) |
pdf |
| 106 |  | Fox:Ebox(Forkhead,bHLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-2 | -5.890e+00 | 0.0101 | 109.0 | 3.63% | 58.7 | 2.35% | motif file (matrix) |
pdf |
| 107 |  | SPCH(bHLH)/Seedling-SPCH-ChIP-Seq(GSE57497)/Homer | 1e-2 | -5.777e+00 | 0.0112 | 104.0 | 3.47% | 55.2 | 2.21% | motif file (matrix) |
pdf |
| 108 |  | Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer | 1e-2 | -5.766e+00 | 0.0112 | 136.0 | 4.53% | 77.1 | 3.08% | motif file (matrix) |
pdf |
| 109 |  | SeqBias: CA-repeat | 1e-2 | -5.531e+00 | 0.0141 | 687.0 | 22.90% | 498.6 | 19.93% | motif file (matrix) |
pdf |
| 110 |  | FOXA1(Forkhead)/MCF7-FOXA1-ChIP-Seq(GSE26831)/Homer | 1e-2 | -5.440e+00 | 0.0153 | 127.0 | 4.23% | 72.6 | 2.90% | motif file (matrix) |
pdf |
| 111 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-2 | -5.411e+00 | 0.0156 | 158.0 | 5.27% | 94.5 | 3.78% | motif file (matrix) |
pdf |
| 112 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-2 | -5.348e+00 | 0.0164 | 19.0 | 0.63% | 4.7 | 0.19% | motif file (matrix) |
pdf |
| 113 |  | Tgif2(Homeobox)/mES-Tgif2-ChIP-Seq(GSE55404)/Homer | 1e-2 | -5.020e+00 | 0.0226 | 538.0 | 17.93% | 385.0 | 15.39% | motif file (matrix) |
pdf |
| 114 |  | HOXA2(Homeobox)/mES-Hoxa2-ChIP-Seq(Donaldson_et_al.)/Homer | 1e-2 | -4.903e+00 | 0.0252 | 22.0 | 0.73% | 6.7 | 0.27% | motif file (matrix) |
pdf |



































































































































































































































